Abstract
Camellia chrysanthoides H. T. Chang and Camellia achrysantha H. T. Chang et S. Y. Liang are two threatened yellow camellia species endemic to southwestern Guangxi, China. Here, we report the complete chloroplast (cp) genomes of C. chrysanthoides and C. achrysantha for the first time. The total cp genome of C. chrysanthoides is 156,959 bp and contains a large single-copy (LSC, 86,564 bp) region, a small single-copy (SSC, 18,267 bp) region, and a pair of inverted repeat (IR, 26,064 bp) regions. The cp genome of C. achrysantha is 156,658 bp and includes an LSC region of 86,249 bp, SSC region of 18,243 bp, and two IR regions of 26,083 bp each. Both C. chrysanthoides and C. achrysantha have 136 genes, including 93 protein-coding genes, 35 tRNA genes, and eight rRNA genes.
Camellia chrysanthoides H. T. Chang and Camellia achrysantha H. T. Chang et S. Y. Liang (family Theaceae) have yellow flowers and are endemic to southwestern Guangxi, China. C. chrysanthoides grows exclusively in acidic soils and has a narrow distribution in Longzhou County (Chang Citation1979). It has been listed as an endangered (EN) species in the Threatened Species List of China's Higher Plants (Qin et al. Citation2017). C. achrysantha grows in calcareous soil and currently only occurs in Tuolu Town, Chongzuo, Guangxi, China. It is a valuable ornamental plant and genetic resource for breeding (Liang Citation1994). C. achrysantha is threatened because of illegal transplanting. Here, we report the complete chloroplast (cp) genomes of C. chrysanthoides and C. achrysantha to provide genetic data to support the conservation and utilization of these two rare species.
Fresh leaves of C. chrysanthoides and C. achrysantha were collected from Daqing Mountain, Longzhou County, Guangxi, China (22.30°N, 106.74°E) and Tuolu Town, Chongzuo, Guangxi, China (22.70°N, 107.72°E), respectively. Voucher specimens of C. chrysanthoides and C. achrysantha were deposited at the Herbarium of Guangxi Institute of Botany (http://ibk.gxib.cn/, Chunrui Lin, [email protected]) under the voucher numbers IBK00430874 and IBK00430873, respectively. Genome sequencing was conducted on the Illumina NovaSeq 6000 Platform. Approximately 5.2 Gb of C. chrysanthoides and 5.3 Gb of C. achrysantha raw reads were obtained. After filtering the low-quality data, ∼5 Gb of clean data were yielded for each species. The trimmed reads were assembled by NOVOPlasty (Dierckxsens et al. Citation2017), and assembled genomes were annotated by PGA (Qu et al. Citation2019) using the complete cp genome of Camellia impressinervis Chang et S. Y. Liang as the reference (GenBank accession number: NC022461).
The complete cp genome sequence of C. chrysanthoides was 156,959 bp in length with a GC content of 37.3%; it had a typical four-conjoined structure: a large single-copy (LSC) region of 86,564 bp, a small single-copy (SSC) region of 18,267 bp, and two inverted repeats (IR) regions, each of 26,064 bp. A total of 136 genes were annotated, including 93 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 35 transfer RNA (tRNA) genes. The complete cp genome of C. achrysantha was similar, with a total length of 156,658 bp (37.3% GC content), including an LSC region of 86,249 bp, SSC region of 18,243 bp, and two IR regions of 26,083 bp each. It contains 136 genes, including 93 PCGs, eight rRNA genes, and 35 tRNA genes.
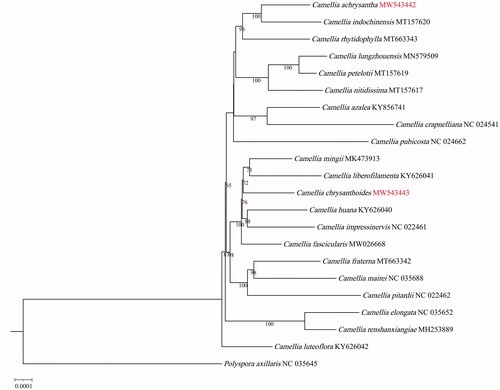
Phylogenomic analysis based on the complete cp genomes was performed using C. chrysanthoides, C. achrysantha, and other cp genome sequences of Camellia. Polyspora axillaris was used as the outgroup. The GenBank accession numbers of the sequences used in this paper are given in . The sequence alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was constructed using IQTREE v1.6.12 (Nguyen et al. Citation2015), the K3Pu + F + 1 model, and 1000 bootstrap replicates. The ML tree showed that C. chrysanthoides was clustered with C. mingii and C. liberofilamenta with 72% bootstrap support (). C. achrysantha was closely related to C. indochinensis with 99% bootstrap support (). Future studies of maternally inherited genetic information in yellow camellias are needed, and rare species of Camellia require protection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession numbers MW543442 and MW543443. The associated BioProject, SRA, and Bio-Sample numbers of C. chrysanthoides are PRJNA695023, SRR13636540, and SAMN17598210, respectively. The associated BioProject, SRA, and Bio-Sample numbers of C. achrysantha are PRJNA695023, SRR13636539, and SAMN17598211, respectively.
Additional information
Funding
References
- Chang HT. 1979. Chrysantha, a section of golden camellias from Cathaysian flora. Act Sci Nat Univ Sunyats. 3:69–74.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome DNA. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liang SY. 1994. Camellia achrysantha, a new species of yellow camellias from Fusui (Guangxi). Guangxi Forestry Sci. 23(1):52–53.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China's higher plants. Biodiv Sci. 25(7):696–744.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.