Abstract
Cladrastis yunchunii X.W.Li et G.S is a plant species belonged to the family Papilionaceae. Cladrastis yunchunii is currently found in broad-leaved forests in the limestone area of Luxi County, Yunnan Province. It is suitable for afforestation and urban greening in limestone areas. In this study, for the first time, we report the complete chloroplast genome sequence of C. yunchunii. We sequenced and assembled the entire chloroplast genome of C. yunchuniii. The chloroplast genome was determined to be 158,250 bp in length. It contained large single-copy (LSC) and small single-copy (SSC) regions of 84,930 bp and 12,664 bp, respectively, which were separated by a pair of inverted repeats (IR) regions of 30,328 bp. The genome contained 132 genes, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The overall GC content of the whole genome is 38.1%, and the corresponding values of the LSC, SSC, and IR regions were 36.4%, 33.6%, and 41.3%, respectively. Phylogenetic analysis suggested that C. yunchunii is closely related to the genus Ormosia in the Papilionaceae.
Cladrastis yunchunii X. W. Li et G. S belongs to the genus Cladrastis in Papilionaceae. Cladrastis yunchunii is currently found in broad-leaved forests in the limestone area of Luxi County (24tly found in b103°30′-104°E), Yunnan Province, China, distributed at altitudes of 1450 m. This species is characterized by petiolule without stipels and inflorescence terminal with white flowers (Li and Fan Citation1994). Additionally, C. yunchunii is currently grown in broad-leaved forests with a small scale distribution characteristics as tall and beautiful trees. It is suitable for afforestation and urban greening in limestone areas (Deng et al. Citation2003). In this study, for the first time, we report the complete chloroplast genome sequence of C. yunchunii.
Until now, the genome research on C. yunchunii has not been revealed, and the complete genome sequence of its chloroplast has not been reported. Complete chloroplast genomes are useful tools for providing the information of species identification, phylogenetic relationships, and germplasm diversity (Wang et al. Citation2019). In this study, we reported the complete genomic sequence of C. yunchunii. And the chloroplast genome will provide a useful resource for C. yunchunii conservation as well as for the phylogenetic studies of Papilionaceae.
The chloroplast genome is a circular DNA molecule (Tonti-Filippini et al. Citation2017). It is highly conserved in plants and an ideal for ecological, evolutionary, and diversity studies (Wicke et al. Citation2011). So far, the complete chloroplast genomes of some species in the family Papilionaceae have been studied and saveddeposited in the GenBank database, e.g. Ormosia hosiei (Zhai et al. Citation2019) and Pterocarpus macrocarpus (Zhang et al. Citation2020). But there is little known about the genome information of C. yunchunii. Therefore, we studied the complete sequence of the chloroplast genome of C. yunchunii and compared it with other known chloroplast genomes to determine phylogenetic relationships among in family Papilionaceae.
The fresh young and healthy leaves of C. yunchunii were collected from Southwest Forestry University Kunming, China (Yunnan, China; geospatial coordinates: 102°45′41″E, 25°04′00″N). The voucher specimens of C. yunchunii were preserved in the herbarium of Southwest Forestry University (Voucher code: SWFU-XW919028). Total genomic DNA was extracted using the Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China). Then DNA were sequenced using the Illumina HiSeq 2000 platform and the compled chloroplast genome were assembled using Velvet and Getorganelle software (Zerbino and Birney Citation2008; Jin et al. Citation2020). Genome annotation was performed with the online annotation tool DOGMA (Dual Organellar GenoMe Annotator, http://dogma.ccbb.utexas.edu/) and program Geneious R8 (Biomatters Ltd, Auckland, New Zealand) (Kearse et al. Citation2012). Finally, the annotated genomic sequence was submitted to GenBank under accession number MW410239.
The complete chloroplast genome of C. yunchunii was 158,250 bp in length, which was contained large single-copy (LSC) and small single-copy (SSC) regions of 84,930 bp and 12,664 bp, respectively, and a pair of inverted repeats (IR) regions of 30,328 bp. The overall GC content of the whole genome was 38.1%, and the corresponding values of the LSC, SSC, and IR regions were 36.4%, 33.6%, and 41.3%, respectively. A total of 132 functional genes were contained in the chloroplast genome, including 87 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes.
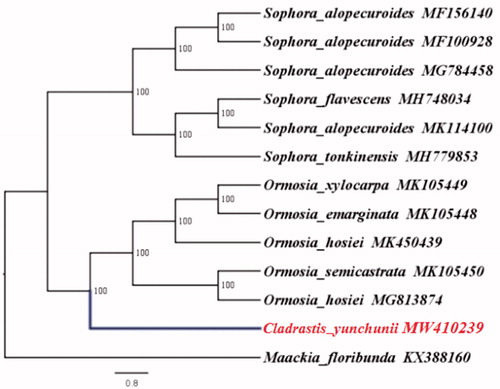
To investigate its taxonomic status, 13 Papilionaceae species plastomes were obtained from the Genbank for phylogenetic analysis, and Maackia floribunda (Papilionaceae) was used as out-group. The 13 complete chloroplast sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). A maximum-likelihood analysis based on the GTR + F+R2 model was performed with iqtree version 1.6.7 using 1000 bootstrap replicates (Nguyen et al. Citation2015). The results revealed that C. yunchunii was closest to Ormosia (). This information was crucial for the correct identification of C. yunchunii and provides valuable genetic resources for the future development of chloroplast derived molecular markers. In conclusion, the complete chloroplast genome of C. yunchunii was decoded for the first time, which will facilitate the species identification, molecular biology, and phylogenetic studies of C. yunchunii. The chloroplast genome will contribute to the research and conservation of C. yunchunii.
Figure 1. Phylogenetic relationships among 13 complete chloroplast genomes (C. yunchunii in this study and 12 previously reported species). Bootstrap values based on 1000 replicates were provided near branches. The ML phylogenetic tree for C. yunchunii is based on the other 12 species (six in Sophora, five in Ormosia, and one in Maackia) chloroplast genomes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no.MW410239. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA687643, SUB8793795, and SAMN17151148 respectively.
Additional information
Funding
References
- Deng LL, Tao SZ, Yuan LF. 2003. Research on tissue culture of Cladrastis yunchunii. J Southwest Forestry Coll. 2:19–25.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–238.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li XW, Fan GS. 1994. A new species of Cladrastis. Plant Res. 4:347–348.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tonti-Filippini J, Nevill PG, Dixon K, Small L. 2017. What can we do with 1000 plastid genomes? Plant J. 90 (4):808–818.
- Wang DW, Chen S, Tang HY, He CZ, Duan AA, Gong HD. 2019. The complete chloroplast genome sequence of Docynia indica (Wall.) Decne. Mitochondrial DNA B Resour. 4(2):3046–3048.
- Wicke S, Schneeweiss GM, Depamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhai DC, Fang Z, Yao Q, Bai XH, Jiang YL, Liu WB, Yang LC, Zhang TT, Zhang Y, Pan J. 2019. Complete chloroplast genome of the wild-type Ormosia hosiei. Mitochondrial DNA Part B. 4(1):1859–1860.
- Zhang J, Li Y, Yuan X, Wang Y. 2020. The complete chloroplast genome sequence of Pterocarpus macrocarpus. Mitochondrial DNA B Resour. 5(1):718–719.
