Abstract
The complete mitochondrial genome of Cochylimorpha cultana (Lederer) (Lepidoptera: Tortricidae) was 15,348 bps in size, and comprised 37 genes, which were 13 PCGs, 22 tRNA genes and two rRNA genes. Most PCGs used the conventional ATN start codon, except for cox1 initiating with CGA. Four genes (cox1, cox2, nad4 and nad5) used single T residue as stop condon. 21 out of 22 tRNAs are folded into the cloverleaf secondary structure, except for trnS1. The phylogenetic analysis based on maximum-likelihood (ML) method revealed that the evolutionary status of C. cultana in Tortricinae at the molecular level, which agrees well with the classical taxonomy.
The leaf roller moth Cochylimorpha cultana belongs to the genus Cochylimorpha Razowski, 1959, which is one of the largest genera in the tribe Cochylini of subfamily Tortricinae. A recent phylogenetic analysis of the tribe placed Cochylimorpha as sister to Eugnosta Hübner, [1825] 1816 (Brown et al. Citation2019). At present, Cochylimorpha comprises 97 species worldwide (Gilligan et al. Citation2018), with greatest species richness in China, Russia and Europe (Sun and Li Citation2013). The larvae of Cochylimorpha utilize mainly Artemisia species (Asteraceae), often feeding on the seeds, stems, and roots (Razowski Citation1987). Numerous species are bound in open, dry biotopes, e.g. sands and various xerotherms; many species are found in the steppes (Razowski Citation2009).
Since the complete mitochondrial genome of Adoxophyes honmai was reported in 2006, there are currently 19 complete mitochondrial genomes of Tortricidae have been published (Lee et al. Citation2006; Son and Kim Citation2011; Zhao et al. Citation2011; Zhu et al. Citation2012; Shi et al. Citation2013; Wu et al. Citation2013; Niu et al. Citation2016; Piper et al. Citation2016; Wu et al. Citation2016; Zhao et al. Citation2016; Fagua et al. Citation2018; Ding et al. Citation2020; Xiang Citation2020). Herein, we sequenced mitochondrial genome of C. cultana, which provided sufficient basis for further analysis of phylogenetic and evolutionary relationship of Tortricidae.
In this study, adult individual of C. cultana was collected from Yanchi County, Ningxia Autonomous Region, China (37.93°N, 107.40°E) in 2017 by light trap. The specimen was identified according to Razowski (Citation1970) and Sun and Li (Citation2013). When the specimen was collected in the field, the three right legs were directly preserved in 95% ethanol and then stored at −20 °C. The remainders of the specimen were deposited as vouchers in the Insect Collection, College of Life Sciences, Dezhou University, Shandong, China (Yinghui Sun, [email protected]), under the accession no. DZU001.
Genomic DNA was extracted from leg muscle using Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, China). The genomic library is established and then used Illumina NovaSeq 6000 platform for high-throughput sequencing performed in Tianjin Novogene Technology Co., Ltd., China. The mitogenome assembly was carried out with the software SPAdes V.3.14.1 (Bankevich et al. Citation2012) and MitoZ V.2.3 (Meng et al. Citation2019). Sequence polish and correctness check were executed with Pilon V.1.23 (Walker et al. Citation2014). MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) was utilized for annotation of the mitogenome (Bernt et al. Citation2013; Cameron Citation2014). Geneious Prime 2020.2.2 was used to compare the homologous gene annotations of other insects and then submitted to NCBI (Kearse et al. Citation2012).
In this study, the circular mitogenome of C. cultana (GenBank accession number: MW413306) was sequenced, assembled and annotated. It was 15,348 bps in size, comprising 37 genes (13 protein-coding genes, 22 tRNA genes, two rRNA genes), in the same gene order as most Ditrysian moth mitogenomes (Park et al. Citation2016; Wang et al. Citation2018; Chen et al. Citation2020). The base composition of C. cultana mitogenome was strongly AT biased (AT 80.8%, CG 19.2%). Most PCGs of C. cultana were using ATN as start codon, while the only exception happened in cox1 which started with putative CGA. With respect to PCG stop codon, the conventional TAA or TAG were most used, except for cox1, cox2, nad4 and nad5 genes which end with a single T residue, where the stop codon is completed by the addition of a poly-A tail to the mRNA. 21 out of 22 tRNAs exhibited the classic cloverleaf structure. TrnS1 as the lone exception possessed a large loop where normally the DHU arm should have formed instead, and additionally, the anticodon of trnS1 was ACU rather than the commonly used GCU. The 372 bp long control region was highly rich in AT content (96.5%).
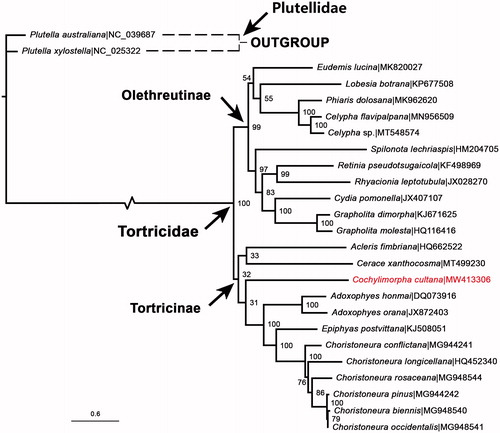
A data set was built based on 13 PCGs of C. cultana plus 24 GenBank sequences from 15 genera, then it was imported into IQ-TREE software V.2.07 (Nguyen et al. Citation2015) to construct a ML tree (), according to the optimal substitution models under the rapid bootstrap algorithm (1000 replicates). The tree generated showed that the monophyly of the two major subfamilies Olethreutinae and Tortricinae was well retained. This study verifies the evolutionary status of C. cultana in Tortricidae at the molecular level, which agreed with the classical taxonomy. The mitochondrial genome would be a significant supplement for the C. cultana genetic background.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW413306. The associated BioProject, BioSample, and SRA numbers are PRJNA730983, SAMN19238733, and SRR14584945, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brown JW, Aarvik L, Heikkil M, Brown R, Mutanen M. 2019. A molecular phylogeny of Cochylina, with confirmation of its relationship to Euliina (Lepidoptera: Tortricidae). Syst Entomol. 45(6):1–15.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):400–411.
- Chen L, Wahlberg N, Liao CQ, Wang CB, Ma FZ, Huang GH. 2020. Fourteen complete mitochondrial genomes of butterflies from the genus lethe (lepidoptera, nymphalidae, satyrinae) with mitogenome-based phylogenetic analysis - sciencedirect. Genomics. 112 (6):4435–4441.
- Ding JH, Yang Y, Li J. 2020. Complete mitochondrial genome of Cerace xanthocosma and its phylogenetic position in the family Tortricidae. Mitochondrial DNA B Resour. 5(3):2906–2908.
- Fagua G, Condamine FL, Brunet B, Clamens AL, Laroche J, Levesque RC, Cusson M, Sperling FAH. 2018. Convergent herbivory on conifers by choristoneura moths after boreal forest formation. Mol Phylogenet Evol. 123:35–43.
- Gilligan TM, Baixeras J, Brown JW. 2018. T@RTS: online world catalogue of the tortricidae (Ver. 4.0). Available from: http://www.tortricidae.com/catalogue.asp.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lee ES, Shin KS, Kim MS, Park H, Cho S, Kim CB. 2006. The mitochondrial genome of the smaller tea tortrix adoxophyes honmai (lepidoptera: tortricidae). Gene. 373:52–57.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Niu FF, Fan XL, Wei SJ. 2016. Complete mitochondrial genome of the Grapholita dimorpha Komai (Lepidoptera: Tortricidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):775–776.
- Park JS, Jeong SY, Kim SS, Kim I. 2016. Complete mitochondrial genome of the gelechioid Hieromantis kurokoi (Lepidoptera: Stathmopodidae). Mitochondrial DNA B Resour. 1(1):285–286.
- Piper MC, Helden MV, Court LN, Tay WT. 2016. Complete mitochondrial genome of the european grapevine moth (egvm) Lobesia botrana (lepidoptera: tortricidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3759–3760.
- Razowski J. 1970. Cochylidae. In: Amsel HG, Gregor F, Reisser H, editors. Microlepidoptera Palaearctica. Vol. 3. IV + p. 528. pp., 161 pls. Wien: Verlag Georg Fromme.
- Razowski J. 1987. The genera of Tortricidae (Lepidoptera). Part I: Palaearctic Chlidanotinae and Tortricinae. Acta Zool Cracov. 30(11):141–355.
- Razowski J. 2009. Tortricidae of the Palaearctic Region, Cochylini. 2: 1–195. Kraków-Bratislava: František Slamka.
- Shi BC, Liu W, Wei SJ. 2013. The complete mitochondrial genome of the codling moth cydia pomonella (lepidoptera: tortricidae). Mitochondrial DNA. 24(1):37–39.
- Son Y, Kim Y. 2011. The complete mitochondrial genome of Grapholita molesta (Lepidoptera: Tortricidae). Ann Entom Soc Amer. 104(4):788–799.
- Sun YH, Li HH. 2013. A brief summary of tribe Cochylini from China (Lepidoptera: Tortricidae: Tortricinae). Hong Kong Entomol Bull. 5(1):13–18.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Wang X, Chen ZM, Gu XS, Wang M, Huang GH, Zwick A. 2018. Phylogenetic relationships among Bombycidae s.l. (Lepidoptera) based on analyses of complete mitochondrial genomes. Syst Entomol. 44:490–498.
- Wu QL, Liu W, Shi BC, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the summer fruit tortrix moth adoxophyes orana (lepidoptera: tortricidae). Mitochondrial DNA. 24(3):214–216.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176.
- Xiang D. 2020. The complete mitochondrial genome of Celypha flavipalpana (Lepidoptera: Tortricidae) from Southeast Tibet of China. Mitochondrial DNA B Resour. 5(3):3547–3548.
- Zhao JL, Wu YP, Su TJ, Jiang GF, Wu CS, Zhu CD. 2016. The complete mitochondrial genome of Acleris fimbriana (Lepidoptera: Tortricidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2200–2202.
- Zhao JL, Zhang YY, Luo AR, Jiang GF, Cameron SL, Zhu CD. 2011. The complete mitochondrial genome of Spilonota lechriaspis meyrick (lepidoptera: tortricidae). Mol Biol Rep. 38(6):3757–3764.
- Zhu JY, Wei SJ, Li QW, Yang S, Li YH. 2012. Mitochondrial genome of the pine tip moth Rhyacionia leptotubula (Lepidoptera: Tortricidae). Mitochondrial DNA. 23(5):376–378.