Abstract
We present the first report of the complete mitochondrial genome of Opsariichthys uncirostris amurensis, which consists of 16,613 base pairs harboring 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and a control region (D-loop). The overall base composition of the complete genome is A (27.15%), C (27.15%), T (26.77%), G (18.92%). The complete mitogenome of O. uncirostris amurensis, which was most closely related to O. bidens in the Bayesian inference tree, provides a better understanding of the phylogeny of the genus Opsariichthys.
Opsariichthys uncirostris amurensis (Berg 1932) (Cypriniformes: Cyprinidae) is a subspecies of O. uncirostris. Opsariichthys uncirostris amurensis lives in most rivers and streams in the Korean Peninsula, except for those flowing to the East Sea of Korea (Kim and Park Citation2002). Opsariichthys uncirostris amurensis has been studied only in phylogenetic analyses of the genus Opsariichthys; studies of O. uncirostris amurensis are lacking (Okazaki et al. Citation2002). This study is the first to report the complete O. uncirostris amurensis mitogenome and provides a better understanding of the phylogenetic relationships in the genus Opsariichthys.
A specimen of O. uncirostris amurensis was collected from Jicheon Stream (36°19'N, 126°55'E) flowing into the Geumgang River in Korea on 18 May 2018, and was preserved in 99.9% ethanol. The specimen was given voucher number SUC-25115 and stored in the fish specimen room of Soonchunhyang University, Korea (Voucher Storage: Soonchunhyang University; Voucher number: SUC-25115; The person in charge of the collection: KR Kim; email: [email protected]). Genomic DNA was extracted from the caudal fin using the Genomic DNA Prep Kit (BioFact, Daejeon, South Korea) and stored in a deep freezer at −80 °C. Genomic DNA was extracted from the caudal fin using the Genomic DNA Prep Kit (BIOFACT, Daejeon, South Korea). The extracted genomic DNA was stored in the specimen room of Soonchunhyang University. To obtain the complete mitochondrial DNA sequence, a DNA library was prepared according to the MGIEasy DNA Library Prep Kit (MGI Tech, Shenzhen, China) manual and subjected to 150 bp paired-end sequencing on the MGISEQ-2000 platform (MGI Tech). The next-generation sequencing raw data were assembled using Geneious 11.0.3, and the assembled mitogenome sequences were annotated using the MITOS web server (Bernt et al. Citation2013). The complete mitogenome sequence of O. uncirostris amurensis was deposited in the National Center for Biotechnology Information GenBank database under the accession number MW355488.
The mitochondrial genome of O. uncirostris amurensis consists of 16,613 bp, harboring 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region (D-loop). Of the 13 PCGs, only the CO1 gene started with GTG, while the remaining 12 started with ATG. Three PCGs (CO2, CO3, and Cytb) contain an incomplete stop codon (T), while 10 PCGs (CO1, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, ATP8, and ATP6) contain a complete stop codon (TAA or TAG). The overall base composition of the O. uncirostris amurensis genome is A (27.15%), C (27.15%), T (26.77%), G (18.92%), with a high AT content of 53.92%. The two rRNAs are 16S (1,693 bp) and 12S (955 bp).
A phylogenetic tree was constructed based on the 13 PCGs using Bayesian inference in MrBayes 3.2.7 (Ronquist et al. Citation2012). For the dataset, we used the GTR + I+G model proposed in jModelTest 2.1.10 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012).
Figure 1. A phylogenetic tree was constructed for the genera Opsariichthys, Zacco, Candidia, Nipponocypris, and Parazacco, with outgroup species and subspecies, using Bayesian inference based on 13 protein-coding genes. The numbers above the nodes are the posterior probabilities of the Bayesian analysis. Scale bars indicate the relative evolutionary distances. The species names are followed by their GenBank accession numbers.
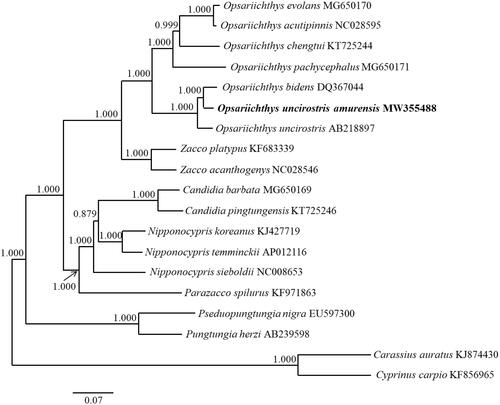
The results indicated that O. uncirostris amurensis is the most closely related to Opsariichthys bidens (Günther 1873). Opsariichthys uncirostris amurensis from the Korean Peninsula was expected to be close to O. uncirostris, which live in Japan, but was more closely related to O. bidens from China (). When the sea level was low, the rivers flowing to the western part of the Korean Peninsula, eastern China, and western Japan were connected to each other through the paleo-Yellow River system. However, it is thought that the river flowing to western Japan was separated geographically due to the sea level rise, and the fish therein evolved into O. uncirostris (Okazaki et al. Citation2002; Takehana et al. Citation2004; Jung et al. Citation2009; Won et al. Citation2020).
For greater insight, a p-distance analysis was performed on O. uncirostris amurensis and species in the genus Opsariichthys. The p-distances between O. uncirostris amurensis and other species of the genus Opsariichthys ranged from 0.137 to 0.142, while O. uncirostris amurensis was 0.054 from O. uncirostris and 0.044 from O bidens. Interestingly, despite being a subspecies of O. uncirostris, O. uncirostris amurensis had a p-distance difference of the similar magnitude as O. bidens. Therefore, p-distance analysis of O. uncirostris amurensis, O. uncirostris, and O. bidens was performed using CO1 sequences in NCBI. The p-distance within O. uncirostris amurensis was in the range 0.002 ∼ 0.007 (HQ536417∼HQ536421, HQ536417∼HQ536421, and MK560566∼MK560568). The p-distance between O. uncirostris amurensis and O. uncirostris was 0.028 and 0.019 ∼ 0.031 for O. bidens (AB218897, LC098444, and MF122583∼MF122595). Therefore, O. uncirostris amurensis and O. uncirostris were recognized as subspecies, although the p-distance differs at the species level. Detailed morphological and molecular phylogenetic analyses are needed. The complete mitogenomes of O. uncirostris amurensis, O. bidens, and O. uncirostris will provide a better understanding of the phylogeny of this species and subspecies.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW355488. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA687317, SRX9720471 and SAMN17141024, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Jung J, Song KH, Lee E, Kim W. 2009. Mitochondrial genetic variations in fishes of the genus Carassius from South Korea: proximity to northern China rather than Japanese Islands? Hydrobiologia. 635. 635(1):95–105.
- Kim IS, Park JY. 2002. Freshwater fishes of Korea. Seoul: Kyohak Press; p. 182–183.
- Okazaki T, Jeon SR, Kitagawa T. 2002. Genetic differentiation of piscivorous chub (genus Opsariichthys) in Japan, Korea and Russia. Zoolog Sci. 19(5):601–610.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Takehana Y, Uchiyama S, Matsuda M, Jeon SR, Sakaizumi M. 2004. Geographic variation and diversity of the cytochrome b gene in wild populations of medaka (Oryzias latipes) from Korea and China. Zoolog Sci. 21(4):483–491.
- Won H, Jeon HB, Suk HY. 2020. Evidence of an ancient connectivity and biogeodispersal of a bitterling species, Rhodeus notatus, across the Korean Peninsula. Sci Rep. 10(1):1–13.
