Abstract
Paphiopedilum parishii (Rchb. f.) Stein is an endangered and rare species with highly ornamental value. In this study, we report the complete chloroplast genome of P. parishii using the Illumina sequencing data. The total genome of P. parishii is 154,689 bp in length and the GC content is 35.9%, with a pair of inverted repeats (IRs) regions of 32,690 bp each, a large single-copy region (LSC) of 86,863 bp and a small single-copy region (SSC) of 2,446 bp. The chloroplast genome encoded 127 genes, including 82 protein-coding genes (CDS), 8 rRNA genes and 37 tRNA genes. The phylogenetic tree showed that P. parishii was clustered with other species in Paphiopedilum with strong support based on the complete chloroplast genome.
Paphiopedilum parishii (Rchb. f.) Stein grows on tree trunks or forks in broad-leaved forests at an altitude of 1,000-1,100 m in China, Laos, Myanmar and Thailand (Chen et al. Citation2009). P. parishii has high ornamental value, since its drooping and curly linear petals have won the favor of many people. However, due to the destruction of habitat, the number and size of its populations are drastically reduced. It has been listed in the Red List of Threatened Species (The International Union for Conservation of Nature, IUCN Citation2015). To better protect P. parishii and provide significant genomic resources in the further study of Orchidaceae, we reported its chloroplast (cp) genome firstly and a neighbour-joining (NJ) phylogenetic tree was constructed for phylogenomic analysis.
We sampled the fresh leaf material from a living individual in the greenhouse of Beijing Forestry University, which was obtained from Guwen village, Jingxi county, Guangxi Zhuang Autonomous Region, China (23.047602°N, 106.316139°E). The specimen was deposited at the herbarium of Plant Biology Department, Beijing Forestry University (Wu Qi. e-mail: [email protected]) under the voucher number BFU-ORCHID-201701. The total genomic DNA was extracted using CTAB method (Doyle and Doyle Citation1987). Then, the next-generation sequencing was performed with an Illumina HiSeq Xten platform at oebiotech (https://www.oebiotech.com/, China). The clean reads were assembled to published cp genomes of Paphiopedilum using the GetOrganelle pipe-line (Jin et al. Citation2018). Finally, we got the complete cp genome of P. parishii and annotated it using PGA (Qu et al. Citation2019). The complete cp genome sequence was submitted to GenBank with the accession number MW528213.
The complete cp genome of P. parishii is 154,689 bp in length, which harbors a typical quadripartite structure with a pair of inverted repeats (IRs) regions of 32,690 bp each, a large single-copy region (LSC) of 86,863 bp and a small single-copy region (SSC) of 2,446 bp. The base composition of the genome is 31.7% A, 18.2% C, 17.7% G, 32.4% T, and the total GC content is 35.9%. The cp genome consists of 127 functional genes, including 82 protein-coding genes (PCGs), 8 rRNA genes and 37 tRNA genes. Among them, 6 tRNA genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC) and 11 protein-coding genes (rps16, atpF, rpoC1, ycf3, clpP, rps12, petB, petD, rpl16, ndhB, rpl2) have introns.
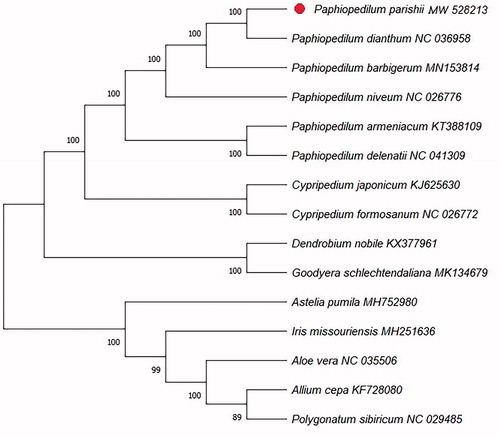
To confirm the phylogenetic position of P. parishii, a molecular phylogenetic tree was constructed with the CDS extracted from the complete cp genome sequence of P. parishii and other 14 complete cp genome sequences (9 species from Orchidaceae and 5 species from other families as outgroup) downloaded from GenBank. All sequences were alignment using the MAFFT (Katoh and Standley Citation2013). The neighbour-joining (NJ) phylogenetic tree was constructed using MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates. The results showed that P. parishii was clustered with other species in Paphiopedilum with strong support (). The data of the complete cp genome of P. parishii provided an essential clue for better understanding the phylogeney and biodiversity of Paphiopedilum.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MW528213. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA741068, SUB9894385, and SAMN19844049 respectively
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Ji ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 25. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- IUCN 2015. The IUCN red list of threatened species. Version 3.1; [accessed 2014 Mar 15]. www.iucnredlist.org.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 4 : 256479. DOI:https://doi.org/10.1101/256479
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Qu XJ, Moore MJ, Li D, Yi TS. 2019. Pga: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.