Abstract
Salix gordejevii is an endemic species in northern China. The analysis of the complete chloroplast genome of S. gordejevii can offer a referential basis for identifying and utilizing Salix germplasm resources. In this study, we obtained chloroplast DNA from S. gordejevii and characterized it. The complete chloroplast genome of S. gordejevii is 155,491 bp in length, comprising a pair of inverted repeats (IR, 27,408 bp), a large single-copy region (LSC, 84,367 bp), and a small single-copy region (SSC, 16,309 bp). We annotated 117 genes in total, including 80 protein-coding genes, 32 tRNA genes, four rRNA genes, and one pseudogene (ycf1). A maximum-likelihood phylogenetic tree was built with MEGA-X and showed that the chloroplast of S. gordejevii has the closest relationship with that of S. magnifica compared to the other reported Salix genomes.
The chloroplast is the core organelle in photosynthesis. Due to its high conservation, the chloroplast genome is widely utilized in species identification and phylogenetic analyses (Szymon et al. Citation2016). The sequencing and analysis of the chloroplast genome are also of great significance in improving plant traits and photosynthetic efficiency (Chen and Liu Citation2008). Salix gordejevii, which belongs to the genus Salix in the family Salicaceae, is widespread in eastern Inner Mongolia, western Liaoning, Gansu, and the Ningxia Hui Autonomous Region, China (Liu Citation1984). It is an important tree species for afforestation and desertification control in the northern region of China due to its strong drought resistance (Cao et al. Citation2018). Additionally, S. gordejevii has become the preferred biological sand barrier for sandstorm source control projects in the dune areas of Beijing-Tianjin (Ma et al. Citation2019). Currently, the complete chloroplast genome sequence of S. gordejevii remains to be sequenced and analyzed, and it can serve as a referential basis to assess the phylogenetic relationships among 24 previously reported Salix species and reconstruct reticulate evolution. In this study, we report the chloroplast genome of S. gordejevii and reconstruct a phylogenetic tree for further analysis.
Fresh leaves of S. gordejevii were collected from the Ningxia Hui Autonomous Region (37.736692°N, 107.348077°E). The voucher specimen (voucher number: NXHL2017002) was deposited at the herbarium of Nanjing Forestry University (https://linxue.njfu.edu.cn/, Xiaoping Li, [email protected]). A modified CTAB method was adapted for the extraction of genomic DNA (Doyle and Doyle Citation1987). DNA fragment libraries were constructed with an average insert size of 300 bp and then sequenced using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA). After removing flanking sequences and low-quality sequences with Fastp (Chen et al. Citation2018), 5.1 GB of high-quality clean reads were assembled with NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome was annotated using GeSeq (Tillich et al. Citation2017) (https://chlorobox.mpimp-golm.mpg.de/geseq.html) with the chloroplast genome of S. dasyclados as a reference (GenBank accession number: MT551160), and the annotated genome was corrected by manual curation. The annotated cpDNA sequence of S. gordejevii was uploaded to GenBank (GenBank accession number: MW562004).
The annotation results indicate that the chloroplast genome of S. gordejevii is 155,491 bp and has a typical quadripartite circular structure. Similar to the chloroplast genome of most higher plants, there are two inverted repeat regions (IRB and IRA, 27,408 bp), a large single-copy region (LSC, 84,367 bp), and a small single-copy region (SSC, 16,309 bp). The CG content of the chloroplast genome is 36.7%. In total, 117 genes were annotated, including 80 protein-coding genes, 32 tRNA genes, four rRNA genes, and one pseudogene.
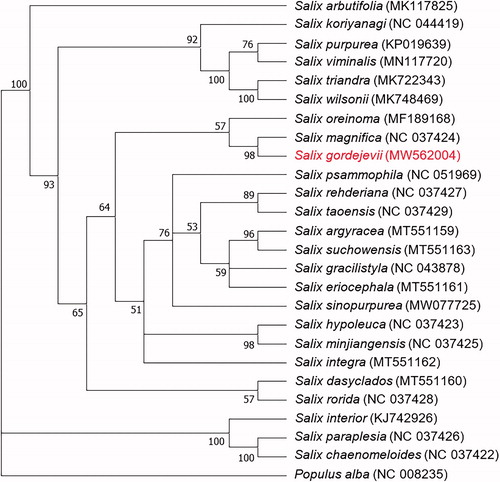
For the phylogenetic analysis of S. gordejevii and other Salix species, a total of 25 chloroplast genomes of Salicaceae were downloaded from NCBI. MAFFT 7 (https://mafft.cbrc.jp/alignment/server/) (Katoh et al. Citation2002) was used to align the sequences. A maximum-likelihood phylogenetic tree (with 1000 bootstrap replicates) was built with a general time reversible (GTR) nucleotide substitution model using the MEGA-X program (Tamura et al. Citation2013), in which Populus alba served as the outgroup. The bootstrap support value (%) is shown next to the nodes. As shown in the phylogenetic tree (), S. gordejevii was most closely related to S. magnifica. The newly annotated plastome of S. gordejevii will be helpful for the phylogenetic identification and utilization of Salix germplasm resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number MW562004. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA734857, SRR14718608, and SAMN19547117 respectively.
Additional information
Funding
References
- Cao GX, Liu XQ, Ji M, Yang YW, Zhang N, Li YX, Li XG, Jia XB, Han SS. 2018. Study on photosynthetic characteristics and light response to drought stress of Salix psammophila and Salix gordejevii. J Inner Mongolia Forestry Sci Technol. 44(04):12–17.
- Chen S, Zhou Y, Chen Y, Jia G. 2018. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics. 34(17):i884–i890.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Liu SE. 1984. Flora Reipublicae Popularis Sinicae. Science Press. 20(2):380.
- Ma SW, Liu GH, Wang L, Lan Q, Xu X. 2019. Effect of drought stress on growth and physiological characteristics of male and female Salix gordejevii cuttings. Acta Bot Boreali-Occidentalia Sin. 39(07):1250–1258.
- Szymon AO, Ewelina Ł, Tomasz K, Tomasz S. 2016. Chloroplasts: state of research and practical applications of plastome sequencing. Planta. 244:517–527.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Chen X, Liu C. 2008. Progress in chloroplast genome analysis. Prog Biochem Biophys. 35:21–28.