Abstract
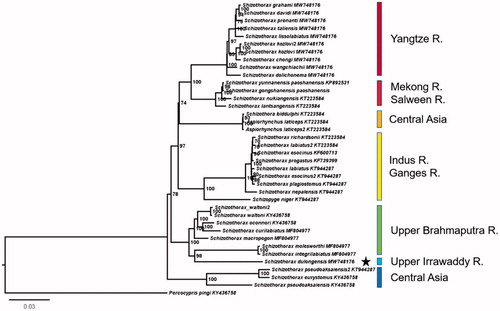
Schizothorax dulongensis Huang (1985) is an endemic species in China, which is mainly distributed in the Irrawaddy River and its tributaries. In this study, the complete mitochondrial genome was sequenced. Its length is 16,579 bp, containing 22 tRNAs, two rRNAs, and 13 PCGs. The phylogenetic results showed that S. dulongensis and these species from Brahmaputra River clustered a clade, and supported the Brahmaputra River and the Irrawaddy River have been reorganized of drainage.
Introduction
Schizothorax fishes are mainly distributed on the Qinghai-Tibet Plateau and surrounding water systems, and they are important organisms for studying the geological evolution of the Qinghai-Tibet Plateau. Most of the complete mitochondrial genome sequences of Schizothorax fishes from other water systems have been reported, but these species from the Irrawaddy River have not been reported, which limits their biogeographical research. Therefore, in this study, the whole mitochondrial genome of S. dulongensis from Irrawaddy River was sequenced and analyzed.
Schizothorax dulongensis belongs to the Cypriniformes, Cyprinidae, Schizothoracinae, and Schizothorax Heckel. The samples were collected from the upper Irrawaddy River (Yunnan, China; 27.843°N, 98.327°E), and a specimen was deposited at Southwest University Museum of Zoology under the voucher number DL-SCH-1-4-20200918 (the contact person is the corresponding author). This mitogenome was submitted to GenBank database with accession No. MW748176.
Material and method
The DNA extraction kit (DC102 FastPure CellTissue DNA Isolation Mini Kit) was used to extract the genomic DNA from S. dulongensis’s pelvic fin. The paired-end (2 × 150 bp) library was constructed and then sequenced on the Illumina Hiseq 5000 platform. The mitochondrial genome was assembled by NOVOPlasty (https://github.com/ndierckx/NOVOPlasty) with Schizothorax davidi (GenBank: KM879227) (Wang et al. Citation2016) as the initial reference, and online tool MITOS (http://mitos2.bioinf.uni-leipzig.de/index.py) was used to annotate the mitochondrial genome. The ML phylogenetic tree using 13 protein-coding gene sequences was constructed by IQ-TREE with default setting and model (Trifinopoulos et al. Citation2016).
Results
The complete mitochondrial genome is 16,579 bp in length, including 22 transfer RNAs (tRNAs) with a total length of 1562 bp, two ribosomal RNAs (rRNAs) with a total length of 2592 bp, and 13 protein-coding genes (PCGs) with a total length of 11,415 bp. The ND6 and eight tRNAs are encoded on the light (L) strain, and the other genes are encoded on the heavy (H) strain. Among PCGs, the start codon of COX1 is GTG, the start codon of other PCGs are ATG; the stop codon of ND2, ATP8 and ND3 are TAG and the rest mostly are TAA as the stop codon. The incomplete stop codon T–– is found in COX2, ND4 and CYTB. The nucleotide composition of the whole mitochondrial genome of S. dulongensis includes 30.21% A, 25.64% T, 17.38% G, and 26.77% C.
The phylogenetic results showed that S. dulongensis and these species from Brahmaputra River clustered a clade, and have clearly differentiated from species in other water systems. This showed the evidence that the Brahmaputra River and the Irrawaddy River had been reorganized of drainage (), and the results supported the conclusions of previous geological studies (Clark et al. Citation2004).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW748176.
Disclosure statement
The authors are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Clark MK, Schoenbohm LM, Royden LH, Whipple KX, Burchfiel BC, Zhang X, Tang W, Wang E, Chen L. 2004. Surface uplift, tectonics, and erosion of eastern Tibet from large-scale drainage patterns. Tectonics. 23(1).
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wang T, Jiao WL, Zhang YP, Lou ZY, Du YY, Yang ZY. 2016. Characterization of the complete mitochondrial genome of Gymnocypris chilianensis (Teleostei: Cypriniformes: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4015–4017.