Abstract
We sequenced and assembled the complete mitochondrial genome of Abscondita cerata from Nankang, Taipei City, Taiwan. The complete mitogenome of A. cerata is 16,964 bp long, and contains 13 protein-coding, 22 tRNA, and two rDNA genes. Nucleotide compositions of the mitogenome of the A. cerata are A: 43.93%, T: 36.74%, C: 11.05%, and G: 8.28%. The AT and GC skewness of the mitogenome sequence are 0.0891 and −0.1434, showing the genome composition skewness toward adenine and cytosine. The clade including all Lampyridae species is well supported. The result indicates that Luciolinae is a monophyletic group but Lampyrinae is not a monophyletic group, as Lamprigera yunnana, which was originally classified into Lampyrinae, is sister to Luciolinae. The genus Lamprigera may share a unique phylogenetic position in Lampyridae. The genus Luciola is a polyphyletic group and the genus Abscondita is a monophyletic group. A. cerata is the sister species to A. chinensis in China. Mitogenomic data from this study will provide useful molecular markers for further studies on the population genetics, speciation, and conservation of endemic species A. cerata in Taiwan.
Lampyridae Rafinesque, 1815, popularly known as fireflies, contains a cosmopolitan group of beetles with approximately 2000 species assigning to 83 genera (Branhm Citation2010). The genus Luciola Laporte is a diverse group containing 283 species (McDermott Citation1966), and many studies based on morphological characters and DNA data revealed that it is not a monophyletic group (Jusoh et al. Citation1975; Ballantyne and Lambkin Citation2001; Chen et al. Citation2019). After the phylogenetic analysis of the species in SE Asian Luciola (sensu lato) by morphological characters, the genus Abscondita was erected and six Luciola species were transferred to it (Ballantyne et al. Citation2013). Subsequently, two new species of Abscondita were described and Luciola pallescens Gorham was transferred to Abscondita, making a total of nine species assigned to the genus (Ballantyne et al. Citation2019). Abscondita cerata (Olivier, 1911) is endemic to Taiwan and it is the only species of Abscondita restricted to one island (Ballantyne et al. Citation2013). Abscondita cerata ranges widely from sea level to 1500 m a.s.l. in Taiwan. Its flight period is mainly from March to May (Chen Citation2003). This is the first report of complete mitochondrial sequences for the species.
Seventeen specimens of A. cerata in this study were collected in Nankang, Taipei City, Taiwan (25°01′40.4″N 121°38′02.6″E) in April 2019. Total genomic DNA of the male specimen was extracted from the thorax of the adult using a ZR Tissue & Insect DNA MicroPrep™ kit (D6015) following the supplier’s instructions. A specimen was deposited at Biodiversity Research Center, Academia Sinica, Taipei, Taiwan (https://www.biodiv.tw/, contact person: T. Y. Wang, [email protected]) under the voucher number 190407NKfly_BM8. The voucher specimen and the other specimens collected in the same site were identified to species level by L. J. Wang based on the references (Chen Citation2003; Ballantyne et al. Citation2013). The complete mitogenome of A. cerata was sequenced using the next-generation sequencing method: the sheared DNA fragments were used for library construction with the MGIEasy DNA Library Prep Kit v1.1. The library was sequenced with the BGISEQ-500RS sequencer by Tri-I Biotech (New Taipei, Taiwan). A total of 9 Gb next-generation sequencing paired-end reads were used to assemble the complete mitogenome sequence. The CLC Genomics Workbench ver.12.0.02 (QIAGEN, Hilden, Germany) was used for sequence quality analysis, data trimming, and de novo assembly by default setting. The assembled contigs were then used to BLAST against the mitogenomes of the congeners A. anceyi (NCBI acc. MH020192) and A. terminalis (NCBI acc. MK292092) to identify the contig with the highest BLAST score and lowest e-value as the mitogenome of A. cerata. The locations of the protein-coding genes, ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) were predicted using MITOS Web Server (Bernt et al. Citation2013) followed by alignment with other mitogenomes of fireflies in the family Lampyridae. The AT and GC skew was calculated according to the following formulas: AT skew=(A – T)/(A + T) and GC skew=(G – C)/(G + C) (Perna and Kocher Citation1995). Maximum-likelihood (ML) analyses were performed using the GTRGAMMA model implemented in RAxML v.8.1.17 (Stamatakis Citation2014). Nodal support confidence was estimated using a fast bootstrapping analysis with 1000 replicates in RAxML with the model GTRCAT.
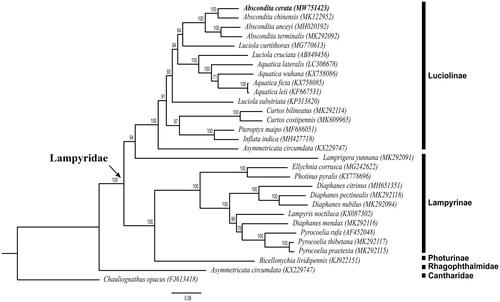
The complete mitogenome of A. cerata is 16,964 bp in length (GenBank accession no. MW751423), including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and one control region. The total nucleotide compositions of the A. cerata mitogenome are 43.93% for A, 36.74% for T, 11.05% for C, and 8.28% for G. The AT and GC skewness of the mitogenome sequence are 0.0891 and −0.1434, showing the genome composition skewness toward adenine and cytosine. The gene rearrangement of the mitogenome in A. cerata is identical to the ancestral inferred insect type (Cameron Citation2014). We reconstructed the phylogenetic relationships including 28 Lampyridae species and two outgroups Rhagophthalmus ohbai (Rhagophthalmidae) and Chauliognathus opacus (Cantharidae) based on 13 mitochondrial protein-coding genes (). Bootstrap values are shown at the branch nodes. The clade consisting of all Lampyridae species is well supported (100%). The phylogenetic tree shows that Lamprigera yunnana, which was originally classified in Lampyrinae (McDermott Citation1966), is sister to Luciolinae. This result is consistent with the molecular phylogenetic result of Chen et al. (Citation2019), indicating that Luciolinae is a monophyletic group while Lampyrinae is not monophyletic. The genus Lamprigera may share a unique phylogenetic position in Lampyridae. The genus Luciola is a polyphyletic group although Luciola substriata have been transferred to the newly erected genus Sclerotia (Ballantyne Citation2016). The clade containing A. cerata (MW751423), A. anceyi (MH020192), A. chinensis (MK122952), and A. terminalis (MK292092) receives absolute support (100%), confirming the genus Abscondita is a monophyletic group. A. cerata is the sister species to A. chinensis in China. More complete mitogenomic data from other Lampyridae species are needed for further studies on the phylogeny of Lampyridae. Mitogenomic data from this study will provide useful molecular markers for further studies on the population genetics, speciation, and conservation of endemic species A. cerata in Taiwan.
Figure 1. Phylogenetic tree of 28 Lampyridae species including Abscondita cerata (in this study, MW751423) and 2 outgroups based on the sequence of mitochondrial 13 protein-coding genes. The tree was reconstructed under the GTRGAMMA model implemented in RAxML v.8.1.17 (Stamatakis Citation2014). Nodal support confidence was estimated using a fast bootstrapping analysis with 1000 replicates in RAxML with the model GTRCAT.
Acknowledgements
We are grateful to Dr. Albert Orr for critical reading the manuscript. Our best thanks go also to Hsieh Jui-Fan, Cho-Yen Lai for DNA data downloading, and Dr. Lesley Ballantyne, Dr. Lars Hendrich for providing relevant references. This work was part of Taiwan Biogenome Project and supported by Academia Sinica, Taiwan.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov under the accession no. MW751423. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA699421, SRR13626958, and SAMN17776035, respectively.
Additional information
Funding
References
- Ballantyne L, Fu XH, Lambkin C, Jeng ML, Faust L, Wijekoon WMCD, Li DQ, Zhu TF. 2013. Studies on South East Asian fireflies: Abscondita, a new genus with details of life history, flashing patterns and behaviour of Abs. chinensis (L.) and Abs. terminalis (Olivier) (Coleoptera: Lampyridae). Zootaxa. 3721(1):1–48.
- Ballantyne L, Lambkin C. 2001. A new firefly, Luciola (Pygoluciola) kinabalua sp. nov. (Coleoptera: Lampyridae), from Malaysia, with observations on a possible copulation clamp. Raffles Bull Zool. 49(2):363–377.
- Ballantyne LA, Lambkin CL, Ho JZ, Jusoh WFA, Nada B, Nak-Eiam S, Thancharoen A, Wattanachaiyingcharoen W, Yiu V. 2019. The Luciolinae of S. E. Asia and the Australopacific region: a revisionary checklist (Coleoptera: Lampyridae) including description of three new genera and 13 new species. Zootaxa. 4687(1):zootaxa.4687.1.1.
- Ballantyne LA, Lambkin CL, Luan X, Boontop Y, Nak-Eiam S, Pimpasalee S, Silalom S, Thancharoen A. 2016. Further studies on South Eastern Asian Luciolinae: 1. Sclerotia Ballantyne, a new genus of fireflies with back swimming larvae 2. Triangulara Pimpasalee, a new genus from Thailand (Coleoptera: Lampyridae). Zootaxa. 4170(2):201–249.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Branhm MA. 2010. Lampyridae Latreille, 1817, pp. 141–149. In: Leschen RAB, Beutel RG, Lawrence JF, editors. Coleoptera, beetles. Volume 2: morphology and systematics (Elateroidea, Bostrichiformia, Cucujiformia partim). Berlin, Germany: Walter de Gruyter.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Chen TR. 2003. The fireflies of Taiwan. Taipei: Field Image Publisher.
- Chen X, Dong Z, Liu G, He J, Zhao R, Wang W, Peng Y, Li X. 2019. Phylogenetic analysis provides insights into the evolution of Asian fireflies and adult bioluminescence. Mol Phylogenet Evol. 140:106600.
- Jusoh WFA, Ballantyne L, Chan SH, Wong TW, Yeo D, Nada B, Chan KO. 1975. Molecular systematics of the firefly genus Luciola (Coleoptera: Lampyridae: Luciolinae) with the description of a new Species from Singapore. Biochem Pharmacol. 24(17):1639–1641.
- McDermott FA. 1966. Lampyridae. In: Steel WO, editor. Coleopterorum Catalogus Supplementa. Pars 9. Editio Secunda. S’Gravenhage, The Netherlands: W. Junk; p. 149.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
