Abstract
In this study, we sequenced the complete plastid genome (plastome) of Neolitsea aciculata, an evergreen broad-leaved tree endemic to East Asia, a woody component of East Asian warm-temperate and subtropical forests across China, Korea, and Japan. The plastome of N. aciculata is assembled as a single contig (152,722 bp). A large and a small single copy (93,785 and 18,795 bp, respectively) of the genome are separated by a pair of inverted repeats (20,071 bp). The genome consists of 126 genes, including 80 protein-coding, eight ribosomal RNA, and 36 transfer RNA genes. Two genes in the IR region (ycf1 and ycf2) are pseudogenized. Our phylogenetic analysis revealed the phylogenetic position of N. aciculata in a highly supported clade of the genus Neolitsea along with other two congeners, N. pallens and N. sericea.
The family Lauraceae consists of approximately 50 genera and 2,500–3,000 species worldwide (Chanderbali et al. Citation2001). Neolitsea (Benth.) Merr., a major genus of the family comprises ca. 100 species widely distributed throughout the tropical and subtropical habitats in Asia, including China, Korea, and Japan (Gentry Citation1988; van der Werff Citation2001; Lee Citation2003; Ohba Citation2006; Huang and van der Werff Citation2008). Its high species diversity is of considerable evolutionary importance as a core genus of Laureae, Litsea complex (Li et al. Citation2004, Citation2007). However, their phylogenetic relationships are still poorly understood.
To date, only two Neolitsea plastid genomes (plastomes) have been sequenced. A study on the woody plants, N. pallens (D. Don) Momiyama and H. Hara and N. sericea (Blume) Koidz., has provided powerful phylogenetic utilities, regarding the Litsea complex in particular (Xiao et al. Citation2020). However, given their high species diversity, such deficiency in the plastome sequence resources on the genus level is a hindrance to improving our understanding of the woody plant-related evolutionary processes in tropical and subtropical forests. Neolitsea aciculata (Blume) Koidz. 1918 is a woody plant of East Asian warm-temperate and subtropical forests across China, Korea, and Japan. In this study, we sequenced and characterized a complete plastome from N. aciculata, endemic to East Asia.
N. aciculata samples were collected from the Jeju Island, South Korea (33°18′26.7″N, 126°27′09.9″E). The voucher specimen (Lee-Na200529) was stored in the department of Biology Education, Chonnam National University (BEC: [email protected]). The DNA library was constructed and sequenced using an MGI-seq 2000 platform (LAS, Seoul, Republic of Korea) following the manufacturer’s protocol. It generated 75,078,260 raw reads (150 bp paired-end). The N. aciculata plastome was assembled using NOVOPlasty 4.1 (Dierckxsens et al. Citation2017), using the N. pallens matK gene sequence (Xiao et al. Citation2020; MN428466) as the seed. The assembled plastome was verified using Geneious 11.0.5 (Kearse et al. Citation2012) by reference mapping 381,661 reads, resulting in a coverage of 150×. The annotation was separately performed using Geneious 11.0.5 and manually corrected for the start and stop codons, as well as the intron-exon boundaries. The annotated plastome sequence was deposited in the GenBank (accession number: MW845678). To construct the phylogenetic tree, plastomes of 10 Lauraceae species (Neolitsea, Lindera, and Litsea: two species each, and Cinnamomum, Laurus, Actinodaphne, and Machilus: one species each, respectively) were downloaded from the NCBI database. The alignments were performed using MAFFT (Katoh and Toh Citation2010). The maximum likelihood (ML) analysis was performed with RAxML v.8.0 (Stamatakis Citation2014) using default parameters and 1000 bootstrap replicates. For the RAxML tree, the general time-reversible (GTR) model of nucleotide substation was used with the Gamma model of rate heterogeneity.
Our results showed that the N. aciculata plastome is 152,722 bp long, with two inverted repeat (IR) regions (20,071 bp each) that separate a large single copy (LSC) region (93,785 bp) and a small single copy (SSC) region (18,795 bp). It contains 126 genes, including 80 protein-coding, eight ribosomal RNA, and 36 transfer RNA genes. Two genes in the IR region, ycf1 and ycf2, are pseudogenized as the nucleotide sequences of the 1,383 bp of 3′–ycf1 and 3,162 bp of 5′–ycf2 truncated at the IR boundaries. The N. aciculata plastome is similar in its gene content and order to those of two congeners, N. pallens and N. sericea. The pseudogenized ycf1 and ycf2 are also present in both congeners. The G + C content is overall 39.1%, of which the LSC, the SSC, and IR regions account for 37.9%, 33.9%, and 44.4%, respectively. Thirteen complete genes (three protein-coding, six tRNA, and four rRNA genes) were duplicated in the IR regions. Fifteen genes exhibit a single intron (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, ndhA, trnK-UUU, trnG-UUC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) and three genes have two introns (ycf3, clpP, and rps12).
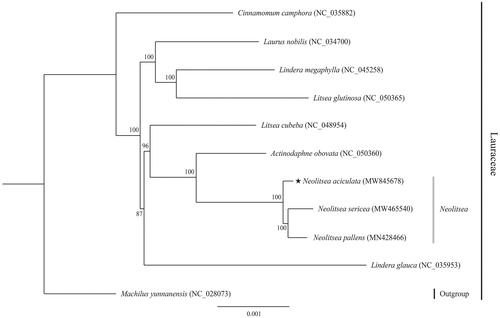
The ML phylogenetic tree shows that N. aciculata is closely related to N. pallens and N. sericea with strong (100%) bootstrap support. The genus Neolitsea was distinguished from the Lindera-related Litsea complex, forming a sister clade with Actinodaphne obovata (). The complete plastome sequenced in this study represents valuable genomic resource data of the Lauraceae, and this new phylogenetic information could be used in future evolutionary studies of the Lauraceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in the NCBI GenBank at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW845678. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726624, SRS8815750, and SAMN18948665, respectively.
Additional information
Funding
References
- Chanderbali AS, van der Werff H, Renner SS. 2001. Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Missouri Bot Gard. 88(1):104–134.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gentry AH. 1988. Changes in plant community diversity and floristic composition on environmental and geographical gradients. Ann Missouri Bot Gard. 75(1):1–34.
- Huang P, van der Werff H. 2008. Neolitsea. Flora of China, Vol. VII. Beijing: Science Press; p. 105–118.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lee TB. 2003. Illustrated flora of Korea, Vol. I. Seoul: Hyangmunsa; p. 408. (in Korean).
- Li J, Christophel DC, Conran JG, Li HW. 2004. Phylogenetic relationships within the ‘core’ Laureae (Litsea complex, Lauraceae) inferred from sequences of the chloroplast gene matK and nuclear ribosomal DNA ITS regions. Plant Syst Evol. 246(1–2):19–34.
- Li L, Li J, Conran JG, Li XW, Li HW. 2007. Phylogeny of Neolitsea (Lauraceae) inferred from Bayesian analysis of nrDNA ITS and ETS sequences. Plant Syst Evol. 269(3–4):203–221.
- Ohba H. 2006. Lauraceae. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan, Vol. IIa. Tokyo: Kodansha; p. 240–253.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- van der Werff H. 2001. An annotated key to the genera of Lauraceae in the Flora Malesiana region. Blumea. 46(1):125–140.
- Xiao TW, Xu Y, Jin L, Liu TJ, Yan HF, Ge XJ. 2020. Conflicting phylogenetic signals in plastomes of the tribe Laureae (Lauraceae). PeerJ. 8:e10155.