ABSTRACT:
The complete chloroplast genome sequence of Thrixspermum amplexicaule was assembled and analyzed in this work. The total chloroplast genome size of T. amplexicaule was 148,124 bp in length, containing a large single-copy (LSC) region of 86,079 bp, a small single-copy (SSC) region of 10,799 bp, and a pair of inverted repeat (IR) regions of 25,623 bp. The GC content of T. amplexicaule was 36.4%. It encoded a total of 120 unique genes, including 75 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The results of phylogenetic analysis strongly supported that all four samples of Thrixspermum are monophyletic and T. amplexicaule was closely related to T. centipeda.
Thrixspermum Loureiro (1790), belonging to Orchidaceae, includes 161 species distributing in the tropical and subtropical regions from Asia to Australia, and only 16 species in China (Pridgeon et al Citation2014; Jin et al. Citation2019). Thrixspermum is close to Phalaenopsis based on the molecular phylogenetic study of Aeridinae (Zou et al. Citation2015). Up to now, only three complete chloroplast genomes of the genus were sequenced and published, including T. centipeda, T. tsii and T. japonicum (Ye et al. Citation2020; Chi et al. Citation2021). Thrixspermum amplexicaule (Blume) Rchb. f. (1867) is a special species that mainly distributing in China, India, Thailand, Vietnam, Malaysia, Indonesia, Philippines and Singapore (Lok et al. Citation2008; Jin et al. Citation2019), and Hainan Island of China is the northernmost boundary of the species in the world. Its flower color often varies from white to purplish and it takes only one day from blooming to falling which makes it difficult to meet pollinator in natural conditions (Jin et al. Citation2019). The species is rare and assessed as a near threatened (NT) species in China (Jin et al. Citation2019). Therefore, developing genomic resources for T. amplexicaule will provide basic information for further study on phylogeny, evolutionary biology and conservation biology.
In this study, the leaf sample of T. amplexicaule was collected from Zuntan Town, Haikou City, Hainan Province of China (110°17'11″E, 19°47'17″N, altitude 44 m). The specimens were deposited at the National Orchid Conservation Center (NOCC, Wen-Hui Rao: [email protected]) and Shanghai Chenshan Botanical Garden (CSH, Bin-Jie Ge: [email protected]) under the voucher number Zheng XL et al. RAD137.
The genomic DNA was extracted from Silica gel dried leaf of T. amplexicaule and sequenced on the Illumina HiSeq platform by Shanghai Origin gene Biotechnology Company (Shanghai, China). DNA extraction, library constructing, sequencing and data filtering were reference in Liu et al. (Citation2019). With the chloroplast genome of T. centipeda (MW057769) as a reference, the paired-end reads were filtered and assembled using GetOrganelle software (Jin et al. Citation2020). The assembled chloroplast genome was annotated with Geneious Prime (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012) and manual adjustment was conducted (Ye et al. Citation2020). The physical map of the chloroplast genome was generated using the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). Finally, we obtained a complete chloroplast genome of T. amplexicaule and submitted to NCBI (Genbank accession MW574621).
The total chloroplast genome size of T. amplexicaule (MW574621) was 148,124 bp in length, containing a large single-copy (LSC) region of 86,079 bp, a small single-copy (SSC) region of 10,799 bp, and a pair of inverted repeats (IR) regions of 25,623 bp. The complete GC content of T. amplexicaule was 36.4%. It encodes a total of 120 unique genes, including 75 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
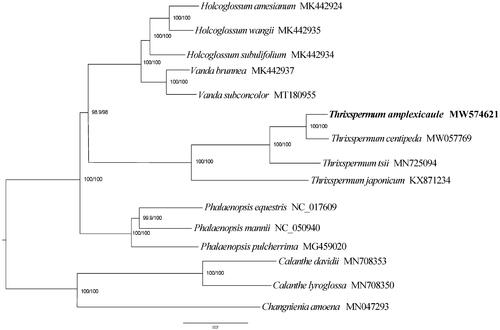
To further understand the intergeneric and intrageneric phylogenetic relationship of Thrixspermum, the matrix of 12 species from Aeridinae and 3 outgroup species (Calanthe lyroglossa, C. davidii and Changnienia amoena) were aligned using MUSCLE version 3.8.31 software (Edgar Citation2004). The phylogenetic tree was constructed using the maximum likelihood method by IQ-TREE, and branch supports with the ultrafast bootstrap (Nguyen et al. Citation2015).
The phylogenetic analysis strongly support that T. amplexicaule was closely related to T. centipeda, and all four samples of Thrixspermum are monophyletic (BS = 100) (). The results were consistent with morphological taxonomy and other phylogenetic study on Orchidaceae (Jin et al. Citation2019; Chi et al. Citation2021; Ye et al. Citation2020; Zou et al. Citation2015).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov) under the accession no. MW574621. The associated ‘BioProject,’ ‘SRA,’ and ‘Bio-Sample’ numbers are PRJNA731595, SRR14618281, and SAMN19291068, respectively.
Additional information
Funding
References
- Chi MW, Liu DK, Zhou CY, Li MH, Lan SR. 2021. The complete plastid genome of Thrixspermum centipeda (Orchidaceae, Aeridinae). Mitochondrial DNA B Resour. 6(3):1245–1246.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Jin XH, Li JW, Ye DP. 2019. Atlas of native orchids in China. Zhenzhou: Henan Science and Technology Press; p. 1018–1079.
- Jin JJ, Yu WB, Yang JB, Song Y, Claude WP, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):231–241.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu DK, Tu XD, Zhang S, Li MH. 2019. The complete plastid genome of Vanda Xichangensis (Orchidaceae, Aeridinae). Mitochondrial DNA B Resour. 4(2):3985–3986.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Lok AFSL, Ibrahim AB, Tan HH, Tan HTW. 2008. Status of Thrixspermum amplexicaule (Bl.) Rchb. f. (Orchidaceae) in Singapore. Nat Singapore. 1:61–64.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2014. Genera Orchidacearum. Volume 6: Epidendroideae (Part 3). Oxford: Oxford University Press.
- Ye BJ, Zhang S, Tu XD, Liu DK, Li MH. 2020. The complete plastid genome of Thrixspermum tsii (Orchidaceae, Aeridinae). Mitochondrial DNA B Resour. 5(1):384–385.
- Zou LH, Huang JX, Zhang GQ, Liu ZJ, Zhuang XY. 2015. A molecular phylogeny of Aeridinae (Orchidaceae: Epidendroideae) inferred from multiple nuclear and chloroplast regions. Mol Phylogenet Evol. 85:247–254.