Abstract
The genus Cuneopsis Simpson, Citation1900 comprises seven valid species, and Cuneopsis celtiformis (Heude, 1874) is the type species of this genus. Previous phylogenetic studies using complete mitochondrial genomes showed that Cuneopsis was not monophyletic, but the result was hampered by incomplete species sampling and lack of the type species of this genus. In this study, we collected C. celtiformis from the type locality and determined its complete maternal mitochondrial genome. This mitogenome is 15,922 bp in length and contains 14 protein-coding genes (including one F-orf), two rRNA genes, 22 tRNA genes, and 1 putative control region. Our mitochondrial phylogenomic analysis confirms that currently recognized genus Cuneopsis is polyphyletic, and C. celtiformis is the closest to C. heudei with high maximum likelihood bootstrap support value. Comprehensive sampling of all Cuneopsis species is needed for phylogenetic analysis to erect new genera in future studies.
Keywords:
The freshwater mussel genus Cuneopsis Simpson, Citation1900, distributed in Asia, comprises seven valid species. Cuneopsis demangei is currently only found in northern Vietnam (Do et al. Citation2018) and the remaining six species are endemic to China (i.e. C. capitatus, C. heudei, C. celtiformis, C. pisciculus, C. rufescens, and C. kiangsiensis) (Simpson Citation1900; Tchang and Li Citation1965). Cuneopsis celtiformis (Heude, 1874) is the type species of this genus, and is mainly distributed in Poyang Lake and Dongting Lake, as well as their connecting rivers (Liu et al. Citation1979). Although previous phylogenetic studies showed that Cuneopsis was not monophyletic (Wu et al. Citation2019; Lopes-Lima et al. Citation2020), the result needs to be further verified by comprehensive sampling of all Cuneopsis species, especially the inclusion of the complete mitogenome of the type species C. celtiformis.
Freshwater mussels have an unusual mode of mitochondrial DNA transmission named Doubly Uniparental Inheritance (DUI). There are two highly differentiated and gender-associated mitogenomes (i.e. maternal and paternal mitogenomes), which vary in sequence and in tissue localization (Breton et al. Citation2007). It has been demonstrated that complete mitogenomes can be effectively used in solving phylogenetic relationships at different taxonomic ranks of freshwater mussels (Lopes-Lima et al. Citation2017; Huang et al. Citation2018; Huang et al. Citation2019; Wu et al. Citation2020). In this study, we first determined and described the complete maternal mitogenome of C. celtiformis, providing useful information for mitogenome comparative analysis and taxonomy of Cuneopsis.
Specimens of C. celtiformis were collected from the Fuhe River (27°56′29″N, 116°26′51″E), Jiangxi Province, China, and were sexed by microscopic examination of gonadal tissues. Total genomic DNA was extracted from the adductor muscle of one live female sample using TIANamp Marine Animals DNA Kit (Tiangen Biotech, Beijing, China). Subsequently, tissues were preserved in anhydrous ethanol, and the voucher specimen (number: 20-NCU-XPWU-CC01; contact Xiao-Chen Huang: [email protected]) was deposited in the Museum of Biology in Nanchang University. The mitogenome was sequenced by primer-walking strategy according to our previous studies (Huang et al. Citation2013; Wu et al. Citation2019). Partial cox1, rrnL and nad1 genes were obtained using universal primers, and then used to design three sets of primers to amplify the complete mitogenome in three long fragments.
SeqMan program (DNAstar) was used to check and assemble the mitogenome. All rRNA and tRNA genes were identified on the MITOS web server (Bernt et al. Citation2013). Protein coding genes (PCGs) were initially identified using the Open Reading Frame Finder (ORF Finder) implemented at the NCBI website (https://www.ncbi.nlm.nih.gov/orffinder/) with invertebrate mitochondrial genetic code. Gene Annotations were further adjusted by comparing BLAST searches with published congeneric mitogenomes.
We downloaded 31 published maternal mitogenomes from the subfamily Unioninae from GenBank, and used Sinosolenaia oleivora, which is from the subfamily Gonideinae, as outgroup. A phylogenetic tree based on 12 PCGs (except atp8 and F-orf) and two rRNA genes was reconstructed under Maximum likelihood (ML) criteria with 10,000 ultrafast bootstrap replicates in IQ-TREE (Nguyen et al. Citation2015). We used ModelFinder (Kalyaanamoorthy et al. Citation2017) to select the best-fit partitioning schemes and models under greedy search with unlinked branch lengths.
The maternal mitogenome of C. celtiformis was 15,922 bp in length and deposited in GenBank under the accession number MW464617. The base composition was A (26.1%), T (38.4%), C (11.7%), and G (23.8%), with the A + T content of 64.5%. It was composed of 14 protein-coding genes (including one F-orf), 22 tRNA genes, 2 rRNA genes, and 1 putative control region (CR). The lengths of the 14 protein-coding genes, in descending order, were nad5, cox1, nad4, cob, nad2, nad1, cox3, atp6, cox2, nad6, nad3, nad4L, F-orf, and atp8. All 22 tRNA genes ranged from 61 to 70 bp, with typical secondary structure. Eleven of the 38 genes were encoded on the H strand, including cox1, cox2, cox3, nad3, nad4, nad4L, nad5, atp6, atp8, tRNAAsp, and tRNAHis. The remaining genes were encoded on the L strand. The location of the F-orf (i.e. between ND2 and tRNAGlu) was the same as that of the other maternal mitogenomes in Unionidae (excluding Gonideinae) (Lopes-Lima et al. Citation2017).
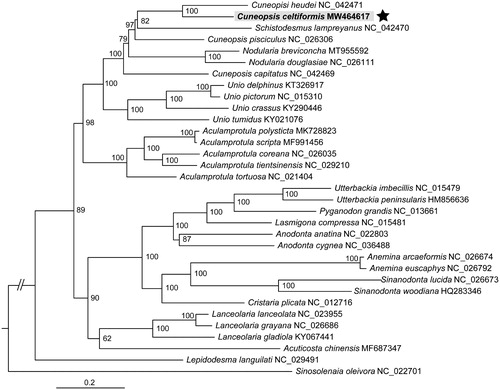
Our phylogenetic tree had bootstrap values ≥95% for most nodes. This ML tree showed that C. celtiformis belonged to the subfamily Unioninae, and was the closest to C. heudei with a high node confidence value (BS = 100) (). Schistodesmus and Nodularia were nested within the currently recognized genus Cuneopsis, respectively. The fact that not all species of Cuneopsis cluster together in the phylogenetic tree based on mitogenome data, which is in accordance with the previous study (Wu et al. Citation2019), confirmed that Cuneopsis is not monophyletic. Comprehensive sampling of all Cuneopsis species is needed for mitochondrial phylogenomics to erect new genera in future studies, together with morphological data.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the Accession no. MW464617.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Breton S, Beaupre HD, Stewart DT, Hoeh WR, Blier PU. 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough? Trends Genet. 23(9):465–474.
- Do VT, Tuan LQ, Bogan AE. 2018. Freshwater mussels (Bivalvia: Unionida) of Vietnam: diversity, distribution, and conservation status. Freshwater Mollusk Biol Conserv. 21(1):1–18.
- Huang X-C, Rong J, Liu Y, Zhang M-H, Wan Y, Ouyang S, Zhou C-H, Wu X-P. 2013. The complete maternally and paternally inherited mitochondrial genomes of the endangered freshwater mussel Solenaia carinatus (Bivalvia: Unionidae) and implications for Unionidae taxonomy. PLOS One. 8(12):e84352.
- Huang X-C, Su J-H, Ouyang J-X, Ouyang S, Zhou C-H, Wu X-P. 2019. Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Mol Phylogenet Evol. 130:45–59.
- Huang X-C, Wu R-W, An C-T, Xie G-L, Su J-H, Ouyang S, Zhou C-H, Wu X-P. 2018. Reclassification of Lamprotula rochechouartii as Margaritifera rochechouartii comb. nov. (Bivalvia: Margaritiferidae) revealed by time-calibrated multi-locus phylogenetic analyses and mitochondrial phylogenomics of Unionoida. Mol Phylogenet Evol. 120:297–306.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Liu Y, Zhang W, Wang Y, Wang E. 1979. Economic fauna of China: freshwater mollusks. Beijing: Science Press.
- Lopes-Lima M, Fonseca MM, Aldridge DC, Bogan AE, Gan HM, Ghamizi M, Sousa R, Teixeira A, Varandas S, Zanatta D, et al. 2017. The first Margaritiferidae male (M-type) mitogenome: mitochondrial gene order as a potential character for determining higher-order phylogeny within Unionida (Bivalvia). J Molluscan Stud. 83(2):249–252.
- Lopes-Lima M, Hattori A, Kondo T, Hee Lee J, Ki Kim S, Shirai A, Hayashi H, Usui T, Sakuma K, Toriya T, et al. 2020. Freshwater mussels (Bivalvia: Unionidae) from the rising sun (Far East Asia): phylogeny, systematics, and distribution. Mol Phylogenet Evol. 146:106755.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Simpson CT. 1900. Synopsis of the Naiades: or pearly freshwater mussels. Washington, DC: Government Printing Office.
- Tchang S, Li S-C. 1965. Bivalves (Mollusca) of the Poyang Lake and surrounding waters, Kiangsi Province, China with description of a new species. Acta Zool Sin. 3:309–317.
- Wu R-W, Liu X-J, Ouyang S, Wu X-P. 2020. Comparative analyses of the complete mitochondrial genomes of three Lamprotula (Bivalvia: Unionidae) species: insight into the shortcomings of mitochondrial DNA for recently diverged species delimitation. Malacologia. 63(1):51–66.
- Wu RW, Liu XJ, Wang S, Roe KJ, Ouyang S, Wu XP. 2019. Analysis of mitochondrial genomes resolves the phylogenetic position of Chinese freshwater mussels (Bivalvia, Unionidae). Zookeys. 812:23–46.