Abstract
We describe the mitochondrial genome sequence of a torrent frog, Amolops jinjiangensis. The mitogenome was extracted and assembled for the first time by restriction site-associated DNA sequencing (RAD-seq). The total length is 17,780 bp, containing 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and one control region. The gene rearrangement was detected as the W-OL-ANCY gene cluster which consisted with several published Amolops mitogenomes. The phylogenetic tree was constructed based on 13 protein-coding genes of A. jinjiangensis and 11 closely related species by Bayesian analyses.
Amolops jinjiangensis, one of the torrent frogs that inhabit rapid-flowing mountain streams or waterfalls, is distributed along the Jinsha River basin in the Hengduan Mountains of southwestern China (Su et al. Citation1986; Frost Citation2021). The previous studies had phylogenetic inferences based on the partial mitochondrial sequences, i.e. COI and Cyt-b, but the phylogenetic relationship and position of A. jinjiangensis were remaining controversial (Lu et al. Citation2014; Lyu et al. Citation2019; Zeng et al. Citation2020). In this study, we identified complete mitochondrial genomes of the A. jinjiangensis by using restriction site-associated DNA sequencing (RAD-seq) to gain additional molecular information and contribute to better understanding of the evolutionary aspects of this frog taxon.
The female adult A. jinjiangensis collected from the type locality, Benzilan Town of Deqin County, Yunnan Province in China (N28°13′57.21″, E99°14′43.38″, 2704 m) in July 2015, and the voucher specimen (CIB-XM6120) was deposited in the Chengdu Institute of Biology, Chinese Academy of Sciences (http://www.cib.ac.cn/, contact Xiaomao Zeng and [email protected]). The muscle tissue isolated from the fresh specimen was preserved in 95% ethanol at −20 °C until use. Genomic DNA was extracted using Genomic DNA Kit (Sangon Biotech, Shanghai) and the single-digest restriction site-associated DNA sequencing (RAD-Seq) library preparation was carried out following the protocol of Baird et al. (Citation2008) by the Novogene (Beijing, China). Briefly, genomic DNA was digested using restriction enzymes EcoRI, and cut-site fragments were sequenced by the Illumina Hiseq-PE150 platform. A total of 9,272,001,300 clean base (bp) was obtained by removing the contaminant sequences and the low-quality regions from raw data. The available clean data of A. jinjiangensis was directly assembled a complete mitogenome by MIRA v4.0.2 and MITObim v1.9.1 (https://github.com/chrishah/MITObim; Hahn et al. Citation2013). We implement the iterations 60 times and take closely related A. granulosus mitogenome (NC_044901.1) as the reference information. Finally, the assembled mitogenome sequence was annotated by using MITOS web server (http://mitos2.bioinf.uni-leipzig.de/index.py; Bernt et al. Citation2013) and the MPI-MP CHLOROBOX tools (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html; Greiner et al. Citation2019).
The whole sequence of the A. jinjiangensis mtDNA was deposited to the GenBank DNA databases under accession number MZ292455. The complete mitogenome of A. jinjiangensis was 17,780 bp in length and it contained the 37 typical genes: two ribosomal RNAs, 22 transfer RNAs (tRNAs), 13 protein-coding genes (PCGs), and one putative control region (D-loop). The length of 12S rRNA was 932 bp, and of 16S rRNA was 1586 bp, while 22 tRNAs ranged from 65 to 73 bp, which were similar to other Amolops species. Overall nucleotides base composition of the complete mtDNA is 28.03% for A, 15.05% for G, 28.87% for C, 28.05% for T, with a higher A + T content (56.08%). The gene rearrangement was detected as the W-OL-ANCY gene cluster in the mitochondrial genome of A. jinjiangensis, which is consistent with five published Amolops mitogenomes including A. mantzorum, A. loloensis, A. tuberodepressus, A. chuanganensis, and A. granulosus, but different from the other three Amolops species, i.e. A. ricketti, A. wuyiensis and A. hongkongensis.
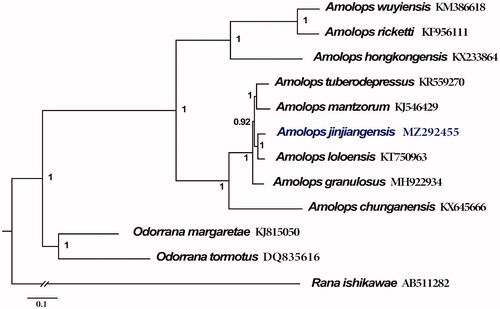
The phylogenetic tree () was constructed based on the 13 PCGs of A. jinjiangensis and 11 closely related species. The best-fit nucleotide substitution models were determined using Partitionfinder 2.1.1 (Lanfear et al. Citation2017). Bayesian analyses were conducted using MrBayes 3.2.7 with the Marko chain Monte Carlo (MCMC) for 20,000,000 generations and 1000 sampled generations (Ronquist et al. Citation2012). The phylogenetic tree indicated that the mitogenome of A. jinjiangensis and A. loloensis clustered together, which supported the phylogenetic inferences estimated by Lyu et al. (Citation2019) and Zeng et al. (Citation2020) with samples also from Deqin County in Yunnan Province, but conflicted with those from Zhongdian County in Yunnan Province (Lu et al. Citation2014). Furthermore, our analyses recovered a sister taxa relationship between A. jinjiangensis + A. loloensis and A. mantzorum + A. tuberodepressus. Although there are no enough mitogenomes of Amolops to analyze phylogenetically, and more information about related species could be useful for a more detailed study of mitogenome evolution and phylogenetic relationships in Amolops.
Figure 1. phylogenetic tree based on the concatenated nucleotide sequences of 13 PCGs from 12 species constructed with Bayesian inference (BI). For the BI tree, Rana ishikawae (AB511282) was used as outgroup. Bayesian posterior probabilities are shown near the nodes. The GenBank accession numbers of all species are shown.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ292455. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA741045, SRR14901856, and SAMN19843588, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3(10):e3376.
- Frost DR. 2021. Amphibian species of the world: an online reference. Version 6.1. New York: American Museum of Natural History. Electronic Database. [accessed 2021 May 16]. Available from: http://research.amnh.org/vz/herpetology/amphibia/index.php.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–64.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Lyu ZT, Zeng ZC, Wan H, Yang JH, Li YL, Pang H, Wang YY. 2019. A new species of Amolops (Anura: Ranidae) from China, with taxonomic comments on A. liangshanensis and Chinese populations of A. marmoratus. Zootaxa. 4609(2):zootaxa.4609.2.3–268.
- Lu B, Bi K, Fu J. 2014. A phylogeographic evaluation of the Amolops mantzorum species group: cryptic species and plateau uplift. Mol Phylogenet Evol. 73:40–52.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Su CY, Yang DT, Li SM. 1986. A new species of Amolops from Hengduan Shan Mountains. Acta Herpetologica Sinica. 5(3):204–206.
- Zeng Z, Liang D, Li J, Lyu Z, Wang Y, Zhang P. 2020. Phylogenetic relationships of the Chinese torrent frogs (Ranidae: Amolops) revealed by phylogenomic analyses of AFLP-Capture data. Mol Phylogenet Evol. 146:106753.
