Abstract
Cynanchum thesioides is a medicinal plant. The complete chloroplast genome sequence of is 158,547 bp in length, contains 131 complete genes, including 85 protein-coding genes (85 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). The overall AT content of cp DNA is 62.1%, the corresponding values of the LSC, SSC, and IR regions are 63.7, 67.7, and 56.5%. Phylogenetic tree shows that C. thesioides was identified as the most divergent among the sequenced species of Cynanchum used.
Cynanchum L. is a large genus in the subfamily Asclepiadoideae (Apocynaceae) with approximately 200 species and a wide distribution throughout the world (Liede and Täuber Citation2002; Endress Citation2018). Cynanchum thesioides (Freyn) K. Schum. (Engler and Anton Citation1895) belonging to genus Cynanchum, is an upright, xerophytic shrub. Cynanchum thesioides has high medicinal value and the plant's fresh juice has been used to treat condyloma acuminatum, plants are rich in rubber and fiber., which can be used as industrial raw materials. plant's fruit edible and seed wool can be used as filling material (Li et al. Citation1995; Zhang et al. Citation2020). However, the chloroplast genome of C. thesioides has not been reported. In this study, we assembled the complete chloroplast genome of C. thesioides, hoping to lay a foundation for further research.
Fresh leaves of C. thesioides were collected from Alxa Youqi (Alxa, Inner Mongolia, China; coordinates: 101°42′E, 39°11′N) and dried with silica gel. The voucher specimen was stored in Sichuan University Herbarium (zhanglei, [email protected]) with the accession number of QTPLJQCHNO0293043. Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987) and a 350-bp library was constructed. This library was sequenced on the Illumina NovaSeq 6000 system with 150 bp paired-end reads. We obtained 10 million high quality pair-end reads for C. thesioides, and after removing the adapters, the remaining reads were used to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017). The complete chloroplasts genome sequence of C. wilfordii was used as a reference. Plann v1.1 (Huang and Cronk Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplasts genome and correct the annotation.
The total plastome length of C. thesioides (MW864598) is 158,547 bp, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 90,848 bp, two inverted repeat (IR) regions of 24,321 bp, and a small single copy (SSC) region of 19,057 bp. The cp genome contains 131 complete genes, including 85 protein-coding genes (85 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes occur in a single copy, while 16 genes occur in double, including all rRNAs (4.5S, 5S, 16S, and 23S rRNA), 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and 5 PCGs (rps7, ndhB, ycf2, rpl2, rpl23). The AT content of the whole plastome is 62.1%, while those of the LSC, SSC, and IR regions are 63.7, 67.7, and 56.5%, respectively.
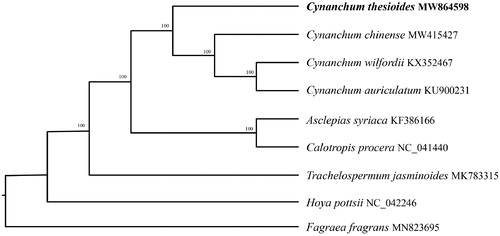
In order to further clarify the phylogenetic position of C. thesioides, plastomes of four representative of Cynanchum species were obtained from NCBI to reconstruct the plastome phylogeny, with Calotropis procera as an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted using RAxML v.8.2.11 (Stamatakis Citation2014) under GTRCAT model with 500 bootstrap replicates () . The phylogenetic tree shows that the species of Cynanchum were clustered together while C. thesioides was identified as the most divergent among the sequenced species of Cynanchum used.
Figure 1. Phylogenetic relationships of Cynanchum species using whole chloroplast genome. GenBank accession numbers: Asclepias syriaca (KF386166), Calotropis procera (NC_041440), Cynanchum auriculatum (KU900231), Cynanchum chinense (MW415427), Cynanchum thesioides (MW864598), Cynanchum wilfordii (KX352467), Fagraea fragrans (MN823695), Hoya pottsii (NC_042246), Trachelospermum jasminoides (MK783315).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW864598. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA672277, SRA: SRS8756897, and SAMN18837310, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Endress ME. 2018. 2582) Proposal to conserve the name Mandevilla against Exothostemon (Apocynaceae: Mesechiteae). Taxon. 67(1):208–209.
- Engler HG, Anton PK. 1895. Die Natürlichen Pflanzenfamilien Nachträge. II: 58–61.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li P, Leeuwenberg A, Middleton DJ. 1995. Flora of China. 16:143-188.
- Liede S, Täuber A. 2002. Circumscription of the genus Cynanchum (Apocynaceae – Asclepiadoideae). System Bot. 27:789–800.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang X, Yang Z, Zhang F, Hao L, Zhao H, Wang C, Long X. 2020. Efect of drought stress on succinic acid biosynthesis in Cynanchum thesioides. J Wuhan Univ. 48:137–145.
