Abstract
Ilex crenata Thunb. is a species of Aquifoliaceae with high ornamental and ecological values. In this study, the complete chloroplast (cp) genome of I. crenata was assembled and characterized through Illumina sequencing data. The entire cp genome of I. crenata was 157,988 bp in length with 37.64% overall GC content, containing a large single-copy (LSC) region of 87,414 bp and a small single-copy (SSC) region of 18,422 bp, which were separated by a pair of 26,076 bp inverted repeat (IR) regions. A total of 135 genes were annotated, including 88 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on 78 conserved protein-coding genes demonstrated that I. crenata is closely related to I. viridis and I. szechwanensis.
Ilex (holly) is the largest woody dioecious genus in the angiosperms containing approximately 700 species within the monogeneric family of Aquifoliaceae (Su et al. Citation2020). The great diversity and adaptability of Ilex plants make them indispensable in gardens and landscapes. Among them, Ilex crenata Thunb. is native to eastern China, Japan and Korea, and has been widely utilized as an ornamental plant for its dense evergreen foliage and various forms (Dirr Citation2009). Many cultivars and hybrids have been bred based on this species (Yang et al. Citation2015). However, the genetic and genomic resources of the species are very limited. Here, we first assembled and characterized the complete chloroplast (cp) genome of I. crenata by Illumina sequencing and bioinformatics analysis, which will contribute to the further studies on the identification and phylogenetic analysis of Ilex species.
I. crenata were planted in Nanjing Botanical Garden, Mem. Sun Yat-sen (E118_83, N32_06), Nanjing, China. The voucher specimen (accession number NBGJIB-Ilex-0037) was stored at the Institute of Botany, Jiangsu Province and Chinese Academy of Science. Fresh leaves were snap-frozen in liquid nitrogen, and the total DNA was extracted from the frozen tissue using the GMS16011.2.1 Kit (Genmed Scientifics Inc., USA). A paired-end library with an insert-size of 350 bp was constructed and sequenced by the Illumina NovaSeq system (Illumina, San Diego, CA) at Novogene company (Tianjin, China). In total, 5607 Mb raw data (5438.4 Mb clean data) were generated. The filtered sequences were used for the de novo genome assembly by the program NOVOPlasty version 3.3 (Dierckxsens et al. Citation2017) and direct-viewing in Geneious R11 (Biomatters Ltd., Auckland, New Zealand). Annotation was performed using GeSeq with further manual correction (Tillich et al. Citation2017). The complete cp genome of I. crenata was deposited in GenBank database (accession number: MW528027).
In general, the whole cp genome of I. crenata was 157,988 bp with 37.64% GC content, including a large single-copy (LSC) region of 87,414 bp, a small single-copy (SSC) region of 18,422 bp and two inverted repeat (IR) regions of 26,076 bp. The cp genome contained 88 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Among them, 8 protein coding genes, 7 tRNA genes and 4 rRNA genes are duplicated in the IR regions. Moreover, fifteen genes contained one intron and three genes (ycf3, clpP and rps12) contained two introns.
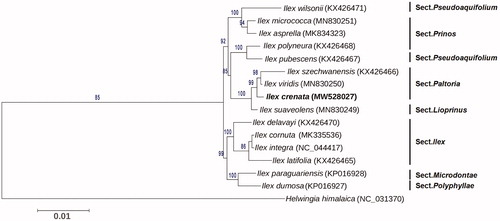
Phylogenetic analysis was performed between I. crenata and other 14 Ilex species, with Helwingia himalaica used as an outgroup (Yao et al. Citation2016; Cascales et al. Citation2017; Park et al. Citation2019; Su et al. Citation2020). All the cp genome sequences other than I. crenata were downloaded from the NCBI database. The 78 common protein-coding genes were extracted and aligned using MAFFT v7.471 (Katoh et al. Citation2019) with default parameters, and then used for tree construction. Phylogenetic tree was constructed by the maximum likelihood (ML) method with the best fit model GTR + I + G using PhyML version 3.0 (Liu et al. Citation2019). The Best-fit model was tested according to the Akaike information criterion (AIC) by jModeltest version 2 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012). The bootstrap values were calculated using 1000 replicates. The phylogenetic tree showed that I. crenata was clustered into the Paltoria section, and closely related to I. viridis and I. szechwanensis (). The complete cp genome sequence of I. crenata will be useful for further analysis on genetic diversity, phylogeny, and molecular breeding.
Figure 1. Maximum likelihood phylogenetic tree based on 78 protein-coding genes of I. crenata and other 15 species. Section names were displayed in the right side of phylogenetic tree. Numbers on the nodes indicated the bootstrap values. Genbank accession number of each species was shown in the brackets after names.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence of I. crenata Thunb. is deposited in Genbank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number MW528027. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA690195, SRR13375982, and SAMN17245922, respectively.
Additional information
Funding
References
- Cascales J, Bracco M, Garberoglio MJ, Poggio L, Gottlieb AM. 2017. Integral phylogenomic approach over Ilex L. species from southern South America. Life. 7(4):47.
- Darriba D, Taboada G, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dirr MA. 2009. Manual of woody landscape plants. 6th ed. Champaign (IL): Stipes Pub. Co.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Katoh K, Rozewicki J, Yamada K. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Liu J, Champer J, Langmüller AM, Liu C, Chung J, Reeves R, Luthra A, Lee YL, Vaughn AH, Clark AG, et al. 2019. Maximum likelihood estimation of fitness components in experimental evolution. Genetics. 211(3):1005–1017.
- Park J, Kim Y, Nam S, Kwon W, Xi H. 2019. The complete chloroplast genome of horned holly, Ilex cornuta Lindl. & Paxton (Aquifoliaceae). Mitochondrial DNA Part B. 4(1):1275–1276.
- Su T, Zhang M, Shan Z, Li X, Zhou B, Wu H, Han M. 2020. Comparative survey of morphological variations and plastid genome sequencing reveals phylogenetic divergence between four endemic Ilex species. Forests. 11(9):964.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:6–11.
- Yang Y, Zhang D, Li Z, Jin X, Dong J. 2015. Immature embryo germination and its micropropagation of Ilex crenata Thunb. horts. 50(5):733–737.
- Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6:28559.
