Abstract
Dendrobium christyanum H.G. Reichenbach (Orchidaceae) is used as a source of the Chinese traditional medicine. Here, we report that the complete chloroplast (cp) genome sequence of D. christyanum is 157972 bp in length with134 genes, of which 114 are unique genes (80 protein-coding genes, 30 tRNAs, and 4 rRNAs). Phylogenetic analysis indicated that D. christyanum was closely related to D. strongylanthum, and D. longicornu. The newly sequenced cp genome will be useful for the phylogenetic and genetic conservation studies of Dendrobium.
The orchid genus Dendrobium Sw. (Epidendroideae, Orchidaceaae) is one of the largest genera in flowering plants, over 1450 species that are mainly distributed in Asia and Oceania (Cribb and Govaerts Citation2005; Xiang et al. Citation2016). Approximately 90 species of Dendrobium are found in China (Zhu et al. Citation2009; Zhang and Feng Citation2018). Most members of this genus have a high ornamental value (Teixeira da Silva et al. Citation2016), and some Dendrobium species have also the medicinal and pharmaceutical values (Teixeira da Silva et al. Citation2015). Dendrobium christyanum H. G. Reichenbach (1882) is an medicinal orchid plant, its short fleshy stems are very suitable for processing into Fengdous (Fengdous, fresh stems of Dendrobium species are processed into the dry products, which have the effects of stimulating saliva, tonifying stomach, and relieving the cough, etc.) (Bao et al. Citation2005). In this study, we sequenced and assembled the complete chloroplast (cp) genome sequence of D. christyanum, and assessed phylogenetic position within Dendrobium, and it could provide useful genomic resources for future studies on identification, phylogeny and evolution of Dendrobium species.
The plants of D. christyanum were collected from Jiangcheng County, Puer City, Yunnan Province, China (101° 51′44″E, 22°35′6″N) and grow in the green house in the Kunming Institute of Botany, Chinese Academy of Sciences. A specimen was deposited at the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN, http://www.kun.ac.cn/, contact person: Feng-Ping Zhang, email: [email protected]) under the voucher number 1511669). Fresh leaves of D. christyanum were harvested for the total genomic DNA extraction using the modified CTAB procedure (Doyle and Doyle Citation1987) and sequenced on Illumina Novaseq 6000 platform (Illumina, San Diego, CA, USA). The complete chloroplast genome was assembled with SPAdes (Bankevich et al. Citation2012, version: 3.13.0) referencing cp genome of closely related Dendrobium species and the genome was annotated with CpGAVAS2 (Liu et al. Citation2012). Finally, we submitted the complete cp genome sequence to the GenBank with accession number MZ241112.
The cp genome sequence of D. christyanum is 157972 bp in length, which presented a typical quadripartite structure, containing a large single-copy region (LSC, 87,717 bp), a small single-copy region (SSC, 18,313 bp), and two inverted repeat regions (IRA and IRB, 25,971 bp). Besides, the cp encoded 134 genes, of which 114 are unique genes (80 protein-coding genes, 30 tRNAs, and 4 rRNAs). In addition, the overall GC content of the whole plastome is 37.23%, whereas the corresponding values of the LSC, SSC, and IRs regions are 34.92, 30.74 and 43.42%, respectively.
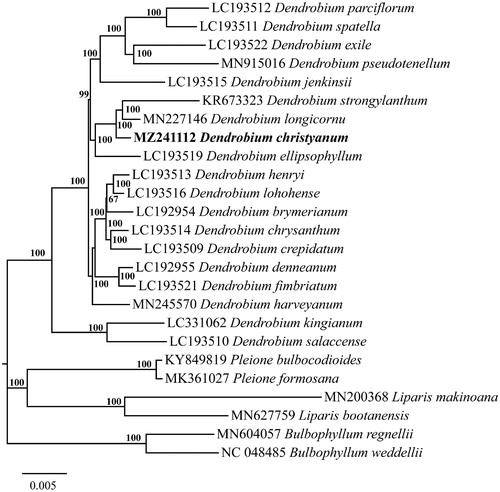
To detect its phylogenetic position, the sequences of D. christyanum and other 18 Dendrobium species (downloaded from NCBI GenBank) were aligned by MAFFT v7.450 software (Katoh and Standley Citation2013), and two Pleione species, two Bulbophyllum species, two Liparis species were selected as outgroup. The phylogenetic tree was constructed by IQ-tree (v2.1.3, http://www.iqtree.org/) with the maximum-likelihood (ML) method. The model GTR + G + I was selected for ML analyses with 5000 ultrafast bootstraps replicates. As expected, D. christyanum was more closely related to D. strongylanthum, and D. longicornu with 100% bootstrap support (), which was consistent with the topology based on molecular analyses of the ITS of the nuclear ribosomal and plastid DNA (rbcL, matK-trnK, trnH-psbA, and trnL-F) in Chen et al. (Citation2016). This newly reported cp genome of D. christyanum is of great benefit to further investigation on its phylogeny and conservation in Dendrobium.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ241112. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA739017, SRR14859971, and SAMN19769087, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly al‐gorithm and its applications to single‐cell sequencing. J Comput Biol. 19(5):455–477.
- Bao XS, Shun QS, Zhang SH, Jin LB. 2005. Zhongguo Yaoyong Shihu Tuzhi. Shanghai: Shanghai Science and Technological Literature Press.
- Chen SP, Ma L, Lan SR, Li MH. 2016. Morphological and molecular evidence for a new species from China: Dendrobiumzhenghuoense (Epidendroideae; Orchidaceae). Phytotaxa. 275(3):277–286.
- Cribb P, Govaerts R. 2005. Just how many orchids are there? In: Raynal- Roques A, Roguenant A, Prat D, editors. Proceedings of the 18th World Orchid Conference. Dijon: Naturalia Publications.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Teixeira da Silva JA, Jin X, Dobranszki J, Lu J, Wang H, Zotz G, Cardoso JC, Zeng S. 2016. Advances in Dendrobium molecular research: applications in genetic variation, identification and breeding. Mol Phylogenet Evol. 95:196–216.
- Teixeira da Silva JA, Tsavkelova EA, Ng TB, Parthibhan S, Dobranszki J, Cardoso JC, Rao MV, Zeng S. 2015. Asymbiotic in vitro seed propagation of Dendrobium. Plant Cell Rep. 34(10):1685–1706.
- Xiang XG, Mi XC, Zhou HL, Li JW, Chung SW, Li DZ, Huang WC, Jin WT, Li ZY, Huang LQ, et al. 2016. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J Biogeogr. 43(7):1310–1323.
- Zhang TM, Feng DQ. 2018. Collection of Dendrobium in China. Beijing: China Medical Science Press.
- Zhu GH, Ji ZH, Wood JJ, Wood HP. 2009. Dendrobium. In: Wu CY, Raven PH, Hong DY, editors. Flora of China. Beijing: Scientific Press.