Abstract
The mountain dragon, Diploderma vela, is an endemic and protected valley lizard that inhabits the upper Lantsang Valley in West China. In this study, we sequenced the complete mitochondrial genome of a male individuals of D. vela using next-generation sequencing methodologies. The complete mitogenome is 16,432 bp in length and contains one noncoding control regions, 13 protein-coding, 22 transfer RNA and two ribosomal RNA genes. The mitogenome content and structure of D. vela was consistent with the previously published representatives of the family. A Bayesian phylogenetic analysis using the complete mitochondrial genomes of Agamidae fully resolved D. vela in the Draconinae, a result consistent with previous investigations. This study provides bioinformatic data for better understanding the evolution and the phylogenetic history of the mountain dragon.
The mountain dragon, Diploderma vela, is an endemic valley lizard that inhabits elevations about 2300 m along the upper Lantsang valley in West China (Wang et al. Citation2015). Unfortunately, the native plant communities and physical structures of valley habitats are being damaged by human activities, such as agricultural planting, road construction and local townships expansion (Wang et al. 2019a). These activities threaten the conservation of this endemic species. This species was recently listed as one of the second-class protected animals of China in early 2021. Meanwhile, our limited knowledge of D. vela has extremely restricted the establishment of conservation strategies and measures (Wang et al. Citation2019b; Uetz et al. Citation2020). In this study, we describe the complete mitochondrial genome of D. vela obtained from next-generation sequencing methods and analyze its phylogenetic relationships with other members within the Agamidae.
A male specimen (CIB5421290081) of D. vela was collected from the type locality of Quzika of Markam county (29°05'N, 98°36'E), Tibet province, China in July 2018. The specimen was identified based on Wang et al. (Citation2015). The specimen and the liver tissue (fixed with 95% ethanol, −20°C) were deposited in the herpetological collection, Chengdu Institute of Biology, Chinese Academy of Science (http://herpmuseum.cib.ac.cn, Li Jia-Tang, [email protected]). Total genomic DNA was extracted from liver tissue using Trelief Animal Genomic DNA Kit (Tsingke, Beijing, China) following the manufacturer’s instruction with minor modification. The complete mitochondrial DNA sequence was analyzed on an Illumina HiSeq 2000 platform. Genes were assembled and annotated with the SPAdes v3.11.0 (Bankevich et al. Citation2012) and MITOS web server (Bernt et al. Citation2013), respectively. The mitogenome was submitted to GenBank under the accession number MW788326. All sampling activities were conducted in accordance with the Guidelines of Animal Ethics published by the Chengdu Institute of Biology.
The complete mitochondrial genomes of D. vela was 16,432 bp in length, comprising one non-coding control region (CR), 13 protein-coding genes (PCGs), two ribosomal RNA genes, and 22 transfer RNA genes (tRNA), while lacking origin of light-strand replication (OL). The mitogenome base-pair is AT biased (58.8%) with 34.8% for A, 28.1% for C, 13.1% for G and 23.9% for T. Most genes were located on the heavy strand (H-strand) with the exception of ND6 and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, SerUCN, Glu, and Pro). The mean length of tRNA genes was 68 bp, the shortest and the longest were tRNA-Cys gene (54 bp) and tRNA-Leu (75 bp), respectively. The mean length of PCGs was 865 bp, the shortest and the longest were ATP8 gene (162 bp) and ND5 (1779 bp), respectively. Most PCGs initiated with ATG except for ATP8, and ND5, both started with GTG. Seven PCGs terminated with complete stop codons, TAA (ND4L, ND5), AGG(ND1), AGA (COX1), CAC (ATP8), and TAG (ND2, and ND6), while the other six genes ended with the incomplete stop codon, TA/T (COX2, ATP6, COX3, ND3, ND4, and Cytb). The mitogenome content and structure of D. vela was consistent with the previously published representatives of the family (Liu et al. Citation2019; Huang et al. Citation2019; Li et al. Citation2021).
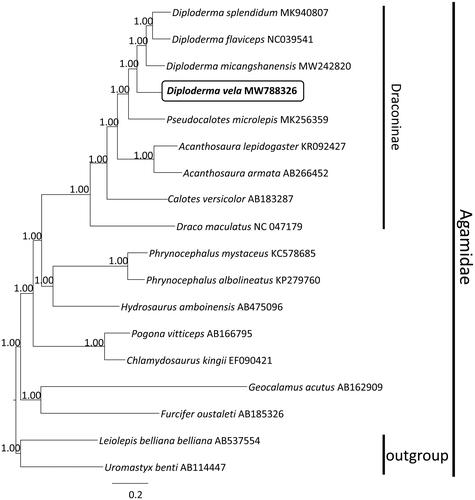
Phylogenetic analysis based on nucleotide sequences of 13 PCGs of D. vela with the other 17 species of Agamidae, both Uromastycinae (Uromastyx benti) and Leiolepidinae (Leiolepis belliana) were designated as outgroups based on published higher-level phylogenetic studies of squamate reptiles (Pyron et al. Citation2013). Bayesian phylogenetic tree using the GTR + I + G substitution model indicated that D. vela was closely related to its congeners, fully resolved in the subfamily Draconinae (PP 1.00) (). The overall phylogenetic relationships among Agamidae were consistent with previous studies (Wang et al. Citation2019a). This study provides a valuable mitogenome resource for better understanding the molecular evolution and phylogenetic relationships of D. vela, and serves as a reference for the establishment of conservation strategies and measures.
Figure 1. Majority rule consensus tree of PCGs of 18 species of Agamidae inferred using MrBayes v.3.2.2 (Ronquist et al. Citation2012) with a GTR + I + G substitution model selected by MrModelTest 2.3 (Nylander Citation2004) under the Akaike information criterion. DNA sequences were aligned in MEGA 7 (Kumar et al. Citation2016). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution. GenBank accession numbers are given with species names.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) with GenBank Accession No. MW788326 (https://www.ncbi.nlm.nih.gov/nuccore/MW788326) and DRYAD (Dryad Digital Repository) with the unique DOI (doi:10.5061/dryad.qjq2bvqgb).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Huang W, Luo H, Luo S, Huang A, Ni Q, Yao Y, Xu H, Zeng B, Li Y, Wei Z, et al. 2019. The complete mitogenome of the splendid japalure Japalura splendida (Squamata, Agamidae). Mitochondrial DNA Part B. 4(2):2641–2642.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li Y, Wang Y, Bai Y, Lv Y, Xiong J. 2021. Mitochondrial genome of Diploderma micangshanense and its implications for phylogeny of the genus Diploderma. Mitochondrial DNA Part B. 6(3):798–802.
- Liu J, Yu J, Zhou M, Yang J. 2019. Complete mitochondrial genome of Japalura flaviceps: deep insights into the phylogeny and gene rearrangements of Agamidae species. Int J Biol Macromol. 125:423–431.
- Nylander JAA. 2004. MrModeltest v2. program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://github.com/nylander.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93 doi:https://doi.org/10.1186/1471-2148-13-93. PMC: 23627680
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Uetz P, Freed P, Hošek J. 2020. The reptile database. [accessed 2019 Nov 9]. http://www.reptile-database.org.
- Wang K, Che J, Lin SM, Deepak V, Aniruddha DR, Jiang K, Jin JQ, Chen HM, Siler CD. 2019a. Multilocus phylogeny and revised classification for mountain dragons of the genus Japalura sl. (Reptilia: Agamidae: Draconinae) from Asia. Zool J Linn Soci. 185(1):246–267.
- Wang K, Jiang K, Pan G, Hou M, Siler CD, Che J. 2015. A new species of Japalura (Squamata: Sauria: Agamidae) from upper Lancang (Mekong) valley of eastern Tibet, China. Asian Herpetol Res. 6(3):159–168.
- Wang K, Jiang K, Ren J, Zou D, Wu J, Che J, Siler CD. 2019b. A new species of Dwarf Japalura sensu lato (Reptilia: Squamata: Agamidae) from the upper Mekong River in Eastern Tibet, China, with notes on morphological variation, distribution, and conservation of two congeners along the same river. Zootaxa. 4544(4):505–522.
