Abstract
The complete mitochondrial genome (mitogenome) of Eoscarta assimilis (Uhler, 1896) was sequenced in the current paper. The total length of the mitogenome is 17,231 bp and it consists of 37 genes including 22 transfer RNA (tRNAs), 13 protein-coding (PCGs) and 2 ribosomal RNA (rRNAs). The 13 PCGs initiated with the start codon ATN, but ND4 started with TTG. All of the PCGs ended with TAA, apart from COX3 which terminated by incomplete TAG. A ML tree based on sequences of 15 complete mitogenomes (13 Cercopidae and 2 outgroup) suggests that E. assimilis is more closely related to the genus Callitettix. The phylogenetic analysis supports the monophyly of the family Cercopidae and the genus Cosmoscarta, and the paraphyly of the subfamily Callitettixinae. This mitogenome information for E. assimilis could facilitate future evolutionary studies to related insects.
The froghopper family Cercopidae is a large group with approximately 1500 species of 150 genera known around the world (Liang and Webb Citation2002). The adults feed on the leaves and stem of a variety of plant and the nymphs may also feed on roots, at or below ground level (Liang and Fletcher Citation2002; Liang Citation2020). Eoscarta assimilis (Uhler, 1896) can be found on the grasses, such as corn and wheat. It is with head and anterior part of pronotum pitchy bromn, scutellum pitchy brown, and mainly distributed in China, Japan, Korea and Russian Maritime Territory (Liang Citation1996).
There are only 12 mitogenomes of froghopper species published and the genus Eoscarta has no record with complete mitogenome in GenBank (https://www.ncbi.nlm.nih.gov/). In this study, we sequenced and assembled the first complete mitogenome of Eoscarta, E. assimilis. A phylogenetic analysis was performed using all known complete mitogenomes of Cercopidae.
The specimens of Eoscarta assimilis were collected from the Huayang National Nature Reserve (107°32′E, 33°36′N, H = 1105m) in Hanzhong City of Shaanxi Province on July 2019, collector is Hu Li, [email protected]. The specimens were immediately preserved in absolute ethanol and frozen at −20 °C and deposited at the Museum of Zoology and Botany, Shaanxi University of Technology, Hanzhong, China (SUHC) with the accession number 2020-14.
Genomic DNA of Eoscarta assimilis was extracted using the TIANamp Genomic DNA kit (Tiangen, Beijing, China). The mitogenome was sequenced using the Illumina NovaSeq 6000 platform, and assembled and annotated with Geneious Prime (Kearse et al. Citation2012). The tRNAs were predicted by ARWEN v1.2 (Laslett and Canback Citation2008), and the rRNAs and control region were identified by alignment with homologous genes of previously determined mitogenomes of Cercopidae.
The whole length of the complete mitogenome of Eoscarta assimilis was 17,231 bp (GenBank no.: MZ047309). This complete mitogenome contains 22 transfer RNA (tRNAs), 13 protein-coding (PCGs), 2 ribosomal RNA (rRNAs) and non-coding regions, of which 23 genes are encoded in J-strand, and the rest of genes are located in N-strand.
The whole nucleotide composition shows significantly A + T bias of 76.7% (45.5% of A; 31.1% of T; 15.0% of C; 8.4% of G). All of the 13 PCGs are initiated with ATN codon, but ND4 used TTG as start codon. Except COX3 ended with TAG codon, others are TAA codon. The length of 22 tRNAs range from 65 (tRNA-Gly) to 73 bp (tRNA-Val). The secondary structure was folded into typical cloverleaf structures, apart from tRNA-Ser1 was missing the D-loop.
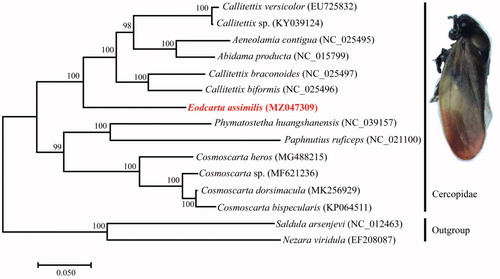
The phylogenetic tree was constructed based on the complete mitogenome sequences from 13 Cercopidae and two outgroups (Saldula arsenjevi and Nezara viridula) (https://www.ncbi.nlm.nih.gov/) using the method of Maximum-Likelihood (ML) with the substitution model and Kimura 2-parameter with 1000 bootstrap replicates using the software MEGA7 (Kumar et al. Citation2016). The result showed Eoscarta assimilis was clustered into Cercopidae and closely related with the genus Callitettix. The monophyly of the family Cercopidae was supported. The monophyly of subfamily Callitettixinae was not supported, which is consistent with Liu et al. (Citation2014) and Yu et al. (Citation2017). The monophyly of the genus Cosmoscarta was controversial (Su et al. Citation2018) while we suggested it was monophyly.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/MZ047309, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA725332, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN18876385 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR14361608.
Figure 1. Phylogenetic tree of Cercopidae based on 15 complete mitogenomes using Maxumum likelihood (ML). Numbers at the nodes represent bootstrap support values based on 1000 replicates and the species in red indicate the new sequence in this study. The adult imaging of Eoscarta assimilis (Uhler, 1896) is showed at upper-right corner.
Additional information
Funding
References
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Liang AP, Fletcher M. 2002. Morphology of the antennal sensilla in four Australian spittlebug species (Hemiptera: Cercopidae) with implications for phylogeny. Aust J Entomol. 41(1):39–44.
- Liang AP, Webb MD. 2002. New taxa and revisionary notes in Rhinaulacini spittlebugs from southern Asia (Homoptera: Cercopidae). J Nat Hist. 36(6):729–756.
- Liang AP. 1996. The spittlebug genus Eoscarta Breddin of China and adjacent areas (Homoptera: Cercopidae). Oriental Insects. 30(1):101–130.
- Liang AP. 2020. A new structure on the frons of male adults of the Asian rice spittlebug Callitettix versicolor (Hemiptera: Auchenorrhyncha: Cercopidae). Zootaxa. 4801(3):591–599.
- Liu J, Bu C, Wipfler B, Liang A. 2014. Comparative analysis of the mitochondrial genomes of Callitettixini Spittlebugs (Hemiptera: Cercopidae) confirms the overall high evolutionary speed of the AT-rich region but reveals the presence of short conservative elements at the tribal level. Plos One. 9(10):e109140.
- Su T, He B, Li K, Liang A. 2018. Comparative analysis of the mitochondrial genomes of oriental spittlebug trible Cosmoscartini: insights into the relationships among closely related taxa. BMC Genomics. 19(1):961.
- Yu PF, Wu GH, Han BY. 2017. Sequencing and analysis of the mitochondrial genome of Kolla paulula (Walker) (Hemiptera: Cicadellidae). J Anhui Agric Univ. 44(5):874–881.
