Abstract
Viscum articulatum is usually used as famous ethno-medicinal plant and popular drink in many provinces of China. In this study, the characterization of the complete chloroplast genome of V. articulatum was analyzed using the Illumina NovaSeq platform. The whole chloroplast genome sequence of V. articulatum is 131,825 including a large single-copy region (LSC, 76,069 bp), a small single-copy region (SSC, 8990 bp), and a pair of repeated regions (IRs, 23,383 bp, each). Further gene annotation in our study revealed the chloroplast genome contains 114 genes, including 36 tRNA genes, 8 rRNA genes and 70 protein-coding genes. A total of 118 simple sequence repeats (SSRs) were identified in the chloroplast genome. Phylogenetic development was analyzed based on V. articulatum with other species of Loranthaceae, the phylogenetic tree in our study revealed that V. articulatum is a lineage independent of other species in genus Viscum.
Viscum articulatum Burm.f. 1768, a parasitic herbaceous perennial plant, is commonly hosted in ancient tea trees of Yunnan province, southwest China (Wang et al. Citation2019). It is also distributed widely in India and Bangladesh (Nag et al. Citation2020). The extracts of this plant contain a wide variety of flavanols, flavonoids, alkaloids, organic acids, and other high molecular weight compounds (Ni et al. Citation2013). V. articulatum is usually used as famous ethno-medicinal plant for the treatment of hemorrhage, pleurisy, gout, heart disease, arthritis, and hypertension (Li et al. Citation2008). It is also used as a popular drink in many provinces of China, which has been developed into expensive commercial products for sale by tea companies (Wang et al. Citation2019). For the above reasons, the pharmacognostical and phytochemical investigation of this plant have been intriguing the interests of researchers (Wang et al. Citation2019; Nag et al. Citation2020; Najafi et al. Citation2010).
In recent years, with the increasing demand for V. articulatum, also because this plant cannot be grown artificially yet, people have been digging and harvesting wild resources more immoderately. Moreover, because this plant grows very slowly, the resources of V. articulatum are significantly decreasing. Considering the chloroplast DNA-based studies can provide invaluable data for studying genetic history and phylogeny, and can also provide important information in conservation and utilization for this species, in this experiment, we collected V. articulatum from Mangjin village, Lancang county of Yunnan province (22°1658′N, 100°0223′E, 1545 m above sea level), China. A voucher specimen (YAB 202007, collector: Yuan Zhang, [email protected]) was deposited at Yunnan Academy of Biodiversity, Southwest Forestry University, Yunnan, China. Then we sequenced, assembled and annotated the accurate chloroplast genome with the method of next-generation sequencing. In this study, we firstly reported a whole mitogenome of V. articulatum (GenBank ID is MW092828), the results will provide more useful information for phylogenetic and evolutionary research of V. articulatum.
For this study, the total genomic DNA of V. articulatum was extracted from fresh stem according to the modified CTAB methods (Doyle and Doyle Citation1987). A genomic shotgun library with an insertion size of 349 bp was constructed, the libraries were sequenced on Illumina NovaSeq platform at Personalbio Biotech (Shanghai, China). The chloroplast genome was assembled using GetOrganelle software version 1.7.1 (Jin et al. Citation2020), and the assembled chloroplast genome was annotated through the program PGA (Plastid Genome Annotator) (Qu et al. Citation2019) with V. coloratum chloroplast genome (GenBank accession number: KY679303) as a reference, assisted with manual correction. The raw sequencing reads used in this study have been deposited in SRA (accession number: SRR12816271), and the annotated chloroplast genome sequence has been deposited into the GenBank (accession number is MW092828).
The complete chloroplast genome of V. articulatum was 131,825 bp and composed of two IRs of 23,383 bp each, which divide a large single copy (LSC) region of 76,069 bp and a small single copy (SSC) region of 8990 bp, the average GC content was 37.50%, with IR regions (43.72%) higher than that in LSC (34.87%) and SSC regions (27.44%). A total of 118 SSR markers ranging from mononucleotide to pentanucleotide repeated motif were identified in the chloroplast genome of V. articulatum. The intron-exon structure analysis indicated that 11 genes have introns, among which atpF, clpP, rpl2, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA have one intron, while ycf3 have two introns. The chloroplast genomes encoded 114 unique genes, including 36 tRNA genes, 8 rRNA genes and 70 protein-coding genes.
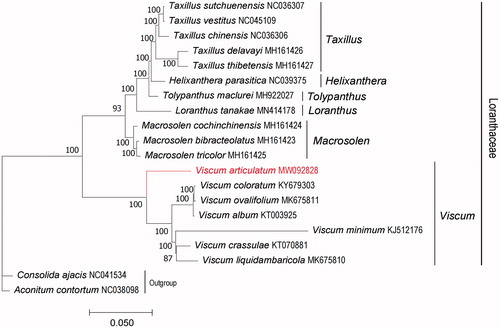
To determine the phylogenetic relationship of V. articulatum, based on complete chloroplast genomes of the other 17 species within the family Loranthaceae, chloroplast genomes were downloaded from NCBI, and we chose Consolida ajacis and Aconitum contortum as outgroups (). All chloroplast genomes were aligned using the program MAFFT v7.471 (Rozewicki et al. Citation2019), and phylogenetic tree (maximum likelihood) constructed by Iqtree software version 1.6.12 (Minh et al. Citation2020) with 1000 bootstrap replicates, best-fitted model has been confirmed is TVM + F + R2 by ModelTest-NG (Darriba et al. Citation2020). The phylogenetic analysis revealed that V. articulatum is a lineage independent of other species in genus Viscum of our study. The study will provide essential data for future research on the phylogenetic and evolutionary relationship of V. articulatum and the family Loranthaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW092828. The associated BioProject, SRA and Bio-Sample numbers are PRJNA668851, SRR12816271, and SAMN16424892 respectively.
Additional information
Funding
References
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Li Y, Zhao YL, Huang N, Zheng YT, Yang YP, Li XL. 2008. Two New phenolic glycosides from Viscum articulatum. Molecules. 13(10):2500–2508.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Nag M, Kar A, Chanda J, Mukherjee PK. 2020. Rp-hplc analysis of methanol extract of Viscum articulatum–a plant from ayurveda. J Ayurveda Integr Med. 11(3):277–280.
- Najafi S, Sadeghinejad B, Deokule SS, Estakhr J. 2010. Phytochemical screening of Bidaria khandalense (sant.) Loranthus capitellatus wall. Viscum articulatum burm.f. and Vitex negundo linn. Res J Pharm Biol Chem Sci. 1(3):388–393.
- Ni TT, Li L, Zhao M, Shao WF, Li JH. 2013. Analysis of flavonols and flavanols in TM Viscum articulatum Burm. Nat Prod Res Develop. 25:484–488.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Rozewicki J, Li SL, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.
- Wang QS, Chen D, Zhang QW, Qin DD, Jiang XH, Li HJ, Fang KX, Cao JX, Wu HL. 2019. Volatile components and nutritional qualities of Viscum articulatum burm.f. parasitic on ancient tea trees. Food Sci Nutr. 7(9):3017–3029.