Abstract
Petunia exserta is an ornamental species on the brink of extinction in the wild. We report here the complete chloroplast genome of P. exserta, which is 156,598 bp in size consisting of a large single-copy region (87,095 bp), a small single-copy region (18,643 bp), and a pair of inverted repeats (25,430 bp for each). The chloroplast (used ‘cp’ hereafter) genome contains 132 genes, including 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes. Phylogenetic analysis demonstrated that P. exserta was most closely related to P. hybrida, and they together were closer to Calibrachoa hybrida than other taxa in the Solanaceae family.
Petunia exserta J.R. Stehm., a recently discovered wild species of the Petunia genus in the Solanaceae (nightshade) family, has an erect growth habit and red star-shaped flowers with exserted styles and anthers. This species was only found in a very small area (about 500 km2) of the Serra do Sudeste region of southern Brazil and is already considered to be near extinction in the wild, with a very limited number of wild plants left in its native habitat (Lorenz-Lemke et al. Citation2006; Segatto et al. Citation2014). Different from the other species of the genus that are pollinated either by bees or by moths, P. exserta is pollinated by hummingbirds, which provide a good system for the genetic study of pollination syndromes (Klahre et al. Citation2011). The speciation process and evolutionary history of P. exserta is intriguing because it can intercross with other Petunia taxa in experimental conditions (Watanabe et al., Citation2001), and some rare cases of natural hybridization between P. axillaris and P. exserta was also found (Lorenz-Lemke et al. Citation2006). Previously, plastid markers were used to analyze the evolutionary relationship of Petunia species, which, however, revealed little genetic differentiation among the species (Ando et al. Citation2005; Kulcheski et al. Citation2006; Lorenz-Lemke et al. Citation2010). Recently, the complete chloroplast genome sequence of P. hybrida, an artificial hybrid between P. integrifolia and P. axillaris, was published (Wong et al. Citation2019). Here we report the complete chloroplast genome of P. exserta, which will provide valuable information for evolutionary studies and hybrid identification in Petunia.
P.exserta was obtained from the Swammerdam Institute for Life Sciences, University of Amsterdam in the Netherlands (gift from prof. Ronald Koes) and stored in the Germplasm Resource Nursery of Ornamental Plants in Guangzhou (N113°20′25″, E23°13′47″). Total genomic DNA was extracted from fresh leaves using the modified 2 × CTAB method (Doyle and Doyle Citation1987). The residual whole plant was processed to a voucher specimen (specimen code SYS-Bore-2020-02-10.3, under the charge of Liu Guofeng, [email protected]) and deposited in Sun Yat-sen University Herbarium. DNA samples were randomly fragmented into ∼400 bp using an ultrasonicator, followed by DNA library construction and paired-end sequencing (2 × 150 bp) on an Illumina Novaseq platform. Approximately 7.58 Gb of raw data was generated and then assembled by GetOrganelle (Jin et al. Citation2020). Sanger sequencing method was used to verify the LSC, SSC and IR junction region which the primers were designed by a online software Primer3 v. 0.4.0 (Koressaar and Remm Citation2007; Untergasser et al. Citation2012) (Table S1). The genome was annotated by using GeSeq (Tillich et al. Citation2017), followed by manual correction and confirmation.
The complete chloroplast genome of P. exserta (GenBank accession no. MT644125) was 156,598 bp in length with total GC content of 37.81%. It contains an 87,095 bp large single-copy (LSC) region, a 18,643 bp small single-copy (SSC) region, and two 25,430 bp inverted repeat (IR) regions. A total of 132 genes were predicted, including 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes, among them all the rRNA genes, 14 tRNA genes, and 12 protein-coding genes locate in the IR regions.
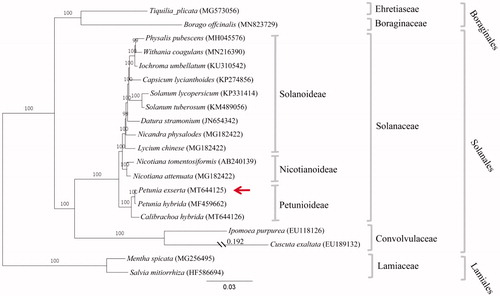
To clarify the evolutionary position of P. exserta, complete chloroplast genomic sequences of 20 species from three orders (Lamiales, Solanales, and Boraginales) were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and then a maximum likelihood (ML) tree was constructed by RAxML version 8 with 1000 bootstrap replicates under the GTRGAMMA substitution model (Stamatakis Citation2014). The phylogenetic analysis showed that P. exserta was clustered together with P. hybrida first, and then closer to Calibrachoa hybrida than other taxa (). Based on the phylogenetic tree, P. exserta, P. hybrida, and C. hybrida form the Petunioideae lineage that was emerged evolutionarily earlier than the Nicotianodeae and Solanoideae lineages, which is consistent with the results reported previously (Wong et al. Citation2019; Liu et al. Citation2020).
Figure 1. Phylogenetic tree based on the complete chloroplast genome sequences of 20 species from Solanales, Lamiales, and Boraginales, with Mentha spicata and Salvia mitiorrhiza as outgroup. The phylogenetic position of P. exserta is indicated by red arrow. Distance was shown for truncated branches. Bootstrap values (1000 replicates) are indicated at nodes. Scale bar: substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT644125 under the accession no. MT644125. And the associated Bioproject, SRA, Bio-sample numbers are PRJNA675663, SRP291795 and SAMN16711698 respectively.
Additional information
Funding
References
- Ando T, Kokubun H, Watanabe H, Tanaka N, Yukawa T, Hashimoto G, Marchesi E, Suaréz E, Basualdo IL. 2005. Phylogenetic analysis of Petunia sensu Jussieu (Solanaceae) using chloroplast DNA RFLP. Ann Bot. 96(2):289–297.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis C, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, Kuhlemeier C. 2011. Pollinator choice in petunia depends on two major genetic loci for floral scent production. Curr Biol. 21(9):730–739.
- Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics. 23(10):1289–1291.
- Kulcheski FR, Muschner VC, Lorenz-Lemke AP, Stehmann JR, Bonatto SL, Salzano FM, Freitas LB. 2006. Molecular phylogenetic analysis of Petunia Juss. (Solanaceae). Genetica. 126(1–2):3–14.
- Liu G, Sun M, Zou P, Zhang W, Ni J. 2020. The complete chloroplast genome sequence of a popular ornamental plant Calibrachoa hybrida (Solanaceae: Petunioideae). Mitochondrial DNA B Resour. 5(3):3374–3393.
- Lorenz-Lemke AP, Mäder G, Muschner VC, Stehmann JR, Bonatto SL, Salzano FM, Freitas LB. 2006. Diversity and natural hybridization in a highly endemic species of Petunia (Solanaceae): a molecular and ecological analysis. Mol Ecol. 15(14):4487–4497.
- Lorenz-Lemke AP, Togni PD, Mäder G, Kriedt RA, Stehmann JR, Salzano FM, Bonatto SL, Freitas LB. 2010. Diversification of plant species in a subtropical region of eastern South American highlands: a phylogeographic perspective on native Petunia (Solanaceae). Mol Ecol. 19(23):5240–5251.
- Segatto ALA, Cazé ALR, Turchetto C, Klahre U, Kuhlemeier C, Bonatto SL, Freitas LB. 2014. Nuclear and plastid markers reveal the persistence of genetic identity: a new perspective on the evolutionary history of Petunia exserta. Mol Phylogenet Evol. 70:504–512.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth CB, Remm M, Rozen GS. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40 (15):e115.
- Watanabe H, Ando T, Tsukamoto T, Hashimoto G, Marchesi E. 2001. Cross compatibility of Petunia exserta with other Petunia taxa. Engei Gakkai Zasshi. 70(1):33–40.
- Wong J, Mudd EA, Hayes A, Day A. 2019. The chloroplast genome sequence of the ornamental plant Petunia hybrida. Mitochondrial DNA Part B. 4(1):249–250.
