Abstract
Oxystophyllum changjiangense has high economic value due to its wide applications in horticultural and medicinal fields. Here, the first chloroplast genome of O. changjiangense was sequenced and reported. The chloroplast genome displayed the typical quadripartite structure containing a pair of inverted repeats (IR), a long single-copy region (LSC), and a short single-copy region (SSC). Total 110 genes were found including 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogeny analysis showed O. changjiangense has a close relationship with Phaius species.
Orchidaceae, one of the largest families in angiosperms, contains approximately 27,000 species distributed in tropical and subtropical regions of the world (Kim et al. Citation2020). Oxystophyllum changjiangense (S. J. Cheng & C. Z. Tang) M. A. Clements 2003 with high ornamental and medicinal value is the only species in Oxystophyllum. Chloroplast genomes of angiosperms were wildly used in molecular taxonomy, species evolution, and authentication. Orchidaceae phylogeny was successfully reconstructed using 76 protein-coding genes from the chloroplast genome (Li et al. Citation2019). Zhu et al. accurately authenticated Dendrobium officinale and its five closely related species based on whole chloroplast genomes (Zhu et al. Citation2018). Here, we sequenced the first chloroplast genome of O. changjiangense. Our aims were to: (1) characterize the structure, content of O. changjiangense chloroplast genome; (2) clarify the phylogenetic position of O. changjiangense.
Individuals of O. changjiangense were sampled from Sanya, Hainan, China (latitude 18.22°, longitude 109.60°) and stored in the greenhouse at Nanjing Normal University (http://www.njnu.edu.cn/, Zhitao Niu, [email protected]) under the voucher number WMT20001. Fresh leaves were collected from O. changjiangense for DNA extraction. Total genomic DNA was isolated using DNeasy Plant Mini Kit (Qiagen, Germany). The DNA sample was deposited at Nanjing Normal University. The quality of the DNA sample was estimated by NanoDrop 8000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and agarose gel electrophoresis. DNA conformed to standard (concentration ≥50 ng/μl, A260/A280 = 1.8-2.0, A260/A230 > 1.8) were used to sequence at Illumina Hiseq4000 platform. Approximately 6 G raw data were generated and to obtain clean data, raw data were filtered by Fast QC. The clean data were trimmed and assembled by CLC Genomics Workbench 8.0 (CLC Bio, Aarhus, Denmark). Gaps and junctions between IR regions and SC regions were confirmed by PCR amplification. Genes were annotated by DOGMA (Wyman et al. Citation2004) and tRNAscan-SE 1.21 (Schattner et al. Citation2005). The complete chloroplast genome of O. changjiangense was submitted to DDBJ (Accession No. LC_579430).
The complete chloroplast genome of O. changjiangense was successfully assembled in this study. The total length of the chloroplast genome was 149,081bp. The chloroplast genome contained four regions including two IR regions (262,11 bp), an LSC region (828,69 bp), and an SSC region (137,90 bp). Total 110 genes were annotated including 76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The total GC content of the chloroplast genome was 37.37% which was similar to other Orchidaceae species (Li et al. Citation2020). The GC content of different regions of the chloroplast genome ranged from 30.26% to 43.27%, and the IR region had the highest GC content.
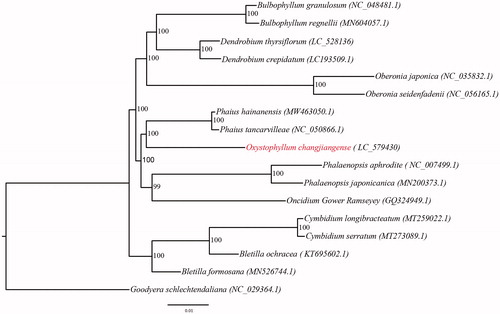
To clarify the phylogenetic position of O. changjiangense, the maximum-likelihood (ML) tree of 17 Orchidaceae species was constructed based on complete chloroplast genomes (). Goodyera schlechtendaliana Rchb. f. was selected as an outgroup. The ML tree was strongly supported (bootstrap supports > 95%). The results showed that Oxystophyllum was sister to Phaius with 100% support, which was consistent with previous studies (Niu et al. Citation2017). We considered that chloroplast genomes have strong power in phylogeny studies.
Figure 1. Maximum-likelihood tree of Orchidaceae species based on the whole chloroplast genome sequences with Goodyera schlechtendaliana as outgroup. Numbers near the nodes represent ML bootstrap values. Oxystophyllum changjiangense is highlighted in red. The contents in parentheses are accession numbers of chloroplast genomes. Oxystophyllum changjiangense is sister to Phaius tancarvilleae with 100% support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number LC_579430. The associated **BioProject**, **SRA**, and **Bio-Sample** numbers are PRJNA669498, SRR12853059, and SAMN16442799 respectively.
Additional information
Funding
References
- Kim YK, Jo S, Cheon SH, Kwak M, Kim YD, Kim KJ. 2020. Plastome evolution and phylogeny of subtribe Aeridinae (Vandeae, Orchidaceae). Mol Phylogenet Evol. 144:106721.
- Li LD, Jiang Y, Liu YY, Niu ZT, Xue QY, Liu W, Ding XY. 2020. The large single-copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Mol Phylogenet E. 69:950–960.
- Li YX, Li ZH, Schuiteman A, Chase MW, Li JW, Huang WC, Hidayat A, Wu SS, Jin XH. 2019. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol Phylogenet Evol. 139:106540.
- Niu ZT, Xue QY, Zhu SY, Sun J, Liu W, Ding XY. 2017. The complete plastome sequences of four orchid species: insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front Plant Sci. 8:715.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhu SY, Niu ZT, Xue QY, Wang H, Xie XZ, Ding XY. 2018. Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm Sin B. 8(6):969–980.
