Abstract
In this study, the complete mitogenome sequence of Korean Boccardiella hamata was determined. This is the first complete mitogenome in the order Spionida. The complete mitogenome of B. hamata is 17,561 bp in length with 12 protein-coding genes (atp8 gene absent), 23 transfer RNAs, 2 ribosomal RNAs, and 1 control region. Interestingly, the gene arrangement of the 12 PCGs of B. hamata is unique, which is very different from that of the other polychaetes currently known. The phylogenetic tree supported the traditional taxonomic position of Spionidae within subclass Sedentaria.
The family Spionidae Grube, 1850 is one of the largest groups of polychaetous annelids that commonly occur in a wide variety of habitats from intertidal to deep-sea and more than 500 nominal species of about 35 genera are currently known (Radashevsky Citation2012). Traditionally, Spionidae is placed in the subclass Sedentaria, which was named after their life habits and the development of the anterior part of the body (Audouin and Milne Edwards Citation1834; Fauchald Citation1977). This is supported in the recent polychaete phylogenetic studies using molecular data (Struck et al. Citation2015; Weigert and Bleidorn Citation2016). Boccardiella hamata (Webster Citation1879) , a relatively widespread species based on current knowledge, is distributed in temperate waters over the northern hemisphere (Kerckhof and Faasse Citation2014). In this study, the complete mitogenome sequence of Korean B. hamata was determined and is the first complete mitogenome in the order Spionida sensu (Rouse and Fauchald Citation1997).
The adult specimens of B. hamata were collected from the mud in crevices between the shells of oysters and their adherent substrates, attached to rocks in intertidal areas at Eurwang-dong, Incheon, South Korea (37°24'59N, 126°24'55E). The voucher specimen was deposited at the National Institute of Biological Resources in South Korea (deposit number: NIBRIV0000886083; Url: https://www.nibr.go.kr; Contact person: Min Seock Do, [email protected]). The mitochondria from a live individual were isolated using Qproteome Mitochondria Isolation Kit (Qiagen, Hilden, Germany). The mitochondrial DNA (mtDNA) was extracted from the mitochondria using a LaboPass Tissue Mini (Cosmo GENETECH, Seoul, South Korea). Whole-genome amplification (WGA) of extracted mtDNA was performed using REPLI-g Mitochondrial DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. The amplified mtDNA was sequenced using Illumina HiSeq 4000 (Macrogen, Seoul, Korea), and assembled by Geneious 8.1.9 (Biomatters Auckland New Zealand) and NovoPlasty v. 2.7.1 (Dierckxsens et al. Citation2017). The complete mitogenome was annotated with the MITOS Webserver (Bernt et al. Citation2013). The extracted mitochondrial DNA was deposited in the DNA collection at the National Institute of Biological Resources in South Korea (deposit number: NIBRGR0000627865). A maximum-likelihood tree was constructed using MEGA X (Kumar et al. Citation2018) under the Tamura-Nei model (Tamura and Nei Citation1993).
The newly determined complete mitogenome of B. hamata (GenBank accession number MW528029) is 17,561 bp in length, containing 12 protein-coding genes (PCGs) (atp8 gene absent), 23 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and one control region (CR). The overall base composition of Korean B. hamata is 29.3% A, 24.1% C, 15.2% G, and 31.4% T, revealing a high A + T content (60.7%). The gene arrangement of the 12 PCGs (cox1-nad6-nad3-cytb- nad4-cox3-atp6-nad5-nad4l-cox2-nad1-nad2) is unique compared to that of the other polychaetes currently available in GenBank.
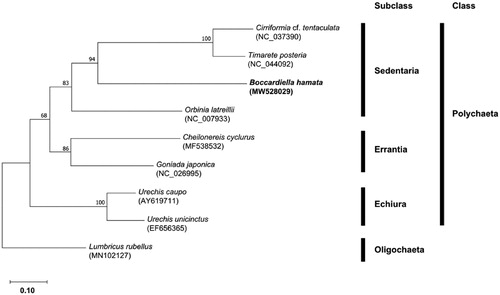
To confirm the molecular phylogenetic relationship, the maximum-likelihood tree was constructed based on the concatenated sequences of 11 protein-coding genes (atp6, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4l, and nad5) from eight polychaetes, including the present sequence. Oligochaete was used as an outgroup (Lumbricus rubellus). As a result, B. hamata belonging to the family Spionidae was clustered with other sedentary polychaetes and formed monophyly (). This supports the traditional taxonomic position of Spionidae within subclass Sedentaria. The newly determined complete mitogenome in the order Spionida will provide useful information for further phylogenetic studies.
Disclosure statement
No potential conflict of interest reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW528029) under the accession no. MW528029. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA748233, SRR15196284, and SAMN20310808, respectively.
Additional information
Funding
References
- Audouin JV, Milne Edwards H. 1834. Recherches pour servir a l'histoire naturelle du littoral de la France, ou Recueil de mémoires sur l'anatomie. La Physiologie, la Classification et les Moeurs Des Animaux de Nos Côtes; Ouvrage Accompagné de Planches Faites D'après Nature. Tome 2, Annélides. Tome 2, premiere part. Paris: Crochard; p. 290.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fauchald K. 1977. The polychaete worms. Definitions and keys to the orders, families and genera. Nat Hist Mus Los Ang. 28:1–188.
- Kerckhof F, Faasse MA. 2014. Boccardia proboscidea and Boccardiella hamata (Polychaeta: Spionidae: Polydorinae), introduced mud worms new for the North Sea and Europe, respectively. Mar Biodivers Rec. 7:e76.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Radashevsky VI. 2012. Spionidae (Annelida) from shallow waters around the British Islands: an identification guide for the NMBAQC Scheme with an overview of spionid morphology and biology. Zootaxa. 3152(1):1–35.
- Rouse GW, Fauchald K. 1997. Cladistics and polychaetes. Zool Scripta. 26(2):139–204.
- Struck TH, Golombek A, Weigert A, Franke FA, Westheide W, Purschke G, Bleidorn C, Halanych KM. 2015. The evolution of annelids reveals two adaptive routes to the interstitial realm. Curr Biol. 25(15):1993–1999.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitution in the control regions of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Webster HE. 1879. The Annelida Chaetopoda of the Virginian coast. Trans Albany Inst. 9:202–269.
- Weigert A, Bleidorn C. 2016. Current status of annelid phylogeny. Org Divers Evol. 16(2):345–362.