Abstract
Polygala crotalarioides Buch.-Ham. ex DC. (Polygalaceae) is a perennial herbaceous plant widely distributed in southwest China. Its chloroplast genome sequence was obtained using Illumina NovaSeq sequencing technology, and then was assembled, annotated, and characterized. The total length of chloroplast genome is 164,269 bp and it contained a large (LCS, 82,697 bp) and a small (SSC, 8083 bp) single copy region, separated by a pair of inverted repeats (IRA/IRB, 36,744 bp). The overall GC content of genome was 36.8% and corresponding values of the LSC, SSC, and IR regions are 34.9%, 29.5%, and 39.7%, respectively. There are 135 genes that were predicted including 89 protein-coding, 38 tRNA, and 8 rRNA genes. The maximum-likelihood (ML) phylogenetic tree reconstructed by IQ-TREE indicated that P. crotalarioides had a strong genetic relationship with P. tenuifolia, P. sibirica, and P. japonica. In summary, the complete chloroplast genome of P. crotalarioides is reported for the first time, which is of great significance for future research on the evolutionary phylogeny and protection of Polygala.
Polygala crotalarioides is a perennial herb classified in the Polygalaceae. It is distributed in southwest and southern China and is an endangered species (Zhang et al. Citation2002). Polygala crotalarioides is considered a rare Chinese herbal medicine used by the Wa people from the Yunnan Province. This plant has excellent biological activities and is widely used in folk to fight fatigue (Xiang and Zhang Citation1995), calm the mind, and tonic the heart. Research shows that P. crotalarioides has anti-oxidative and anti-fatigue effects and improves the body’s ability to adapt to stress (Liu and Ma Citation2014). Analysis of the chloroplast genome will contribute to the bioinformatics and evolutionary history of this species.
Chloroplast genomes can provide reliable data to determine genetic and evolutionary relationships in plants (Kugita et al. Citation2003). So far, complete chloroplast genomes of several species from the Polygalaceae have been studied and deposited in GenBank, including P. japonica (Zuo et al. Citation2021). At present, the chloroplast genome of P. crotalarioides has not been deciphered. In this paper, we assembled and annotated the chloroplast genome of P. crotalarioides using high-throughput sequencing. These data provide a reference for further molecular research on the taxonomy and evolutionary systematic of the Polygalaceae.
Fresh leaves of P. crotalarioides were collected from Shuangjiang County (Yunnan, China; geospatial coordinates: 99.844°E, 23.514°N; Altitude: 1015.9 m). The total genomic DNA was extracted using the Magnetic beads plant genomic DNA preps Kit (Annoroad Biological Technology, Yiwu, China). The herbarium specimen was deposited at the Herbarium of Southwest Forestry University Southwest Forestry University, Kunming, China (http://bbg.swfu.edu.cn/, Shuang-Zhi Li, [email protected]) under the voucher number PC20200910-5.
The Illumina NovaSeq 6000 platform was used for sequencing and the read length was 150 bp paired end. The filtered chloroplast reads were assembled from scratch using GetOrganelle. Then, we selected P. tenuifolia as the reference sequence, using Geneious R8 (Biomatters Ltd, Auckland, New Zealand), assembled, and performed the annotation of the complete chloroplast genome. Finally, the chloroplast DNA sequence and complete annotation were deposited in GenBank under accession number MW543308.1.
The chloroplast genome is 164,268 bp in length and consists of an LSC (82,697 bp) and SSC (8,083 bp), separated by two inverted repeats (36,744 bp). The total GC content in the LSC (34.9%) and SSC (29.5%) was 36.8%, which was lower than that in the IR regions (39.7%). The circular genome contains 135 genes, including 89 protein-coding (PCG), 38 tRNA, and 8 rRNA genes. Among the 135 annotated genes, there are 84 single copy genes and 25 duplicate genes. The protein-coding genes, tRNA and rRNA of P. crotalarioides chloroplast genome are identical in quantity with P. japonica, but they are different in total length, total GC content, etc., which may be caused by differences in species and gene richness (Zuo et al. Citation2021).
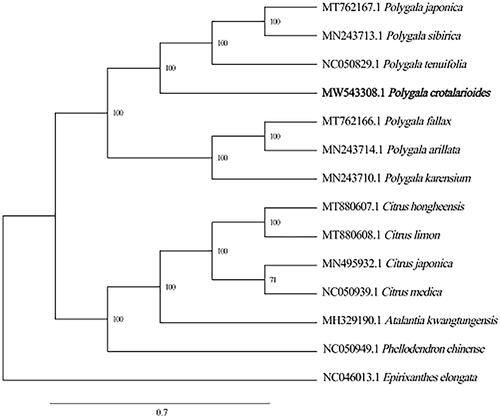
To uncover the phylogenetic relationship of P. crotalarioides in Polygalaceae family, the chloroplast genome sequences of 14 plants including P. crotalarioides were selected to construct the phylogenetic trees. The alignment of all cp genomes was done by the MAFFT version 7 software (Katoh and Standley Citation2013), which was analyzed by IQ-TREE 1.5.5 (Nguyen et al. Citation2015) under the TVM + F+R2 nucleotide substitution model (Kalyaanamoorthy et al. Citation2017). The phylogenetic analysis 1000 bootstrapping replicates was performed based on the maximum likelihood (ML) tree. Phylogenetic analysis showed that Polygala crotalarioides was strongly related to P. tenuifolia, P. sibirica, and P. japonica with 100% bootstrap support (). Among them, the chloroplast structure of P. crotalarioides we studied was highly consistent with that of P. tenuifolia of the same genus previously published (Lee et al. Citation2020). In conclusion, the complete chloroplast genome of P. crotalarioides provides useful DNA data for further biological analysis and the evolutionary phylogenetics of the Polygalaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number MW543308.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA732229, SRR14679146, and SAMN19314347 respectively.
Additional information
Funding
References
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kugita M, Kaneko A, Yamamoto Y, Takeya Y, Matsumoto T, Yoshinaga K. 2003. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res. 31(2):716–721.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Lee D-H, Cho W-B, Park BJ, Kim JD, Byeon JG. 2020. The complete chloroplast genome of Polygala tenuifolia, a critically endangered species in Korea. Mitochondrial DNA Part B. 5(2):1919–1920.
- Liu C, Ma HY. 2014. Overview of traditional Chinese medicine Polygala tenuifolia. Hebei Agricult Sci. 18(5):75–81.
- Xiang BX, Zhang PF. 1995. Investigation on the wa herbal medicine "yamonang" and its resources. Gui Zhou Sci. 013(001):24–28.
- Zhang PX, Duan R, Huang P. 2002. Resources and geographical distribution of medicinal plants of Polygala in China. Res Pract Modern Chin Med. 16(06):42–43.
- Zuo Y, Mao Y, Shang S, Yang G. 2021. The complete chloroplast genome of Polygala japonica houtt. (Polygalaceae), a medicinal plant in China. Mitochondrial DNA Part B. 6(1):239–240.