Abstract
Ophioglossum vulgatum is a rare and ancient fern. In this study, the chloroplast (cp) genome of O. vulgatum was completely sequenced. The genome size is 138,562 bp, which contains a large single-copy (LSC) region with 99,351 bp, a small single-copy (SSC) region with 19,661 bp, and two inverted repeats (IR) regions of 9,775 bp each. Additionally, the overall GC content is 42.14%. It encodes a total 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The Bayesian phylogenetic tree shows that O. vulgatum and O. californicum formed a monophyletic branch. This study can provide a molecular basis for studying the phylogenetic genomics and population variation of Ophioglossaceae.
Ophioglossum vulgatum (Ophioglossaceae) is a rare and ancient fern. It is mainly distributed in the northern hemisphere, generally growing in wet mountains, river banks, and ditches (Zhu et al. Citation2014). The whole plant can be used as medicine (Yang et al. Citation2019). It can clear away heat, diminish inflammation, and treat some cancers (Lu et al. Citation2009). It is known as the ‘Medicine King’ in Taiwan, China (Hu et al. Citation2016). In addition, it has important scientific research value in plant systematics and pteridophyte phylogeny (Zhu et al. Citation2014). The relationships among the branches of Ophioglossaceae were not well solved in previous molecular studies (Zhang, Fan, et al. Citation2020). Obtaining the entire chloroplast genome can provide a molecular basis for studying the phylogenetic genomics and population variation of Ophioglossaceae.
Fresh leaves were sampled from the campus of the South China Agricultural University (SCAU) (E113°20′, N23°9′). The specimen is stored in the Herbarium of South China Agricultural University (SCAUB, specimen code: JHao202105; herbaria acronyms follow Thiers Citation2021, continuously updated). Genomic DNA was extracted using Plant Genomic DNA Kit (Kangweishiji CW0553, China). Illumina Novaseq6000 (Illumina, San Diego, CA) high-throughput sequencing platform was used for sequencing, and the sequencing strategy was PE150 (Pair-End 150). Clean reads with high quality were obtained by filtering the original sequences. A total of 7,587,551 clean reads were generated. The chloroplast (cp) genome sequence was assembled using SPAdes version 3.5.0 (Bankevich et al. Citation2012). SPAdes with multi kmers from 79 to 97, and other paratemers were set default. We used CpGAVAS (Liu et al. Citation2012) and ORFfinder (National Library of Medicine, U.S., National Center for Biotechnology Information, 2004; cited 2021 April 25. Available from: https://www.ncbi.nlm.nih.gov/orffinder/) for cp genome annotation. For the preliminary annotation results, the methods of Blastn and Blastp (National Library of Medicine, U.S., National Center for Biotechnology Information, 2004; cited 2021 April 25. Available from: https://blast.ncbi.nlm.nih.gov/Blast.cgi/) were used to compare and verify the coding proteins and rRNAs of the cp genomes with related species. The annotation of tRNA was carried out by ARWEN (Laslett and Canback Citation2008). If abnormal tRNA occurred, tRNAscan-SE2.0 (Lowe and Chan Citation2016) was used to predict jointly, and finally, tRNAs with unreasonable length and incomplete structure were discarded. Fifteen species were selected to reconstruct the phylogenetic tree. The program MAFFT plugin (selected "-auto" strategy) in PhyloSuite version 1.2.1 (Zhang, Gao, et al. Citation2020) was used to create a multiple sequence alignment of the complete cp genome of O. vulgatum plus 14 other plants, in which the sequences were downloaded from GenBank (Equisetum arvense was selected as outgroup). GTR + F+I + G4 was chosen as the best-fit model according to the Bayesian Information Criterion (BIC). We reconstructed the Bayesian phylogenetic tree with PhyloSuite (2,000,000 generations, Nst = 6, rates = invgamma) ().
Figure 1. The Bayesian phylogenetic tree constructed based on the complete chloroplast genome sequences of 15 species. Equisetum arvense was selected as outgroup. The posterior probability of Bayesian was 1.
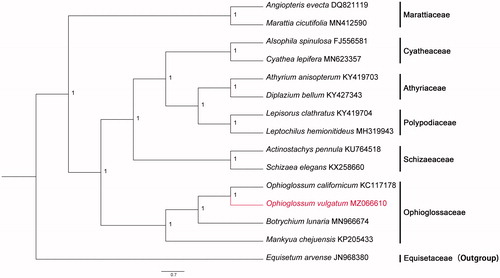
The complete cp genome of O. vulgatum is 138,562 bp in size with circular DNA molecular structure. It contains a large single-copy (LSC) region with 99,351 bp, a small single-copy (SSC) region with 19,661 bp, and two inverted repeats regions (IRs) of 9775 bp each. The overall GC content of this cp genome is 42.14%. The GC contents in LSC, SSC, and IR regions were 40.60%, 38.99%, and 53.14%, respectively. The cp genome encodes a total 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these genes, eight genes (atpF, ndhA, ndhB, rpl2, rpl16, petB, petD, and rpoC1) contain one intron, while two genes (clpP, ycf3) have two introns. Additionally, the ratios of A, T, G, and C are 28.94%, 28.92%, 20.54%, and 21.59%. As shown in the Bayesian tree (), the phylogenetic analysis shows that O. vulgatum and O. californicum formed a monophyletic branch. The cp genome of O. vulgatum can provide valuable genomic information to further phylogenetic relationship of Ophioglossaceae.
Disclosure statement
The authors declare that they do not have any conflict of interest.
Data availability statement
The data that support the findings of this study are available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/), reference number MZ066610. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA739081, SRX11193688, and SAMN19771167, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Hu WC, Meng QY, Wan CX. 2016. Fingerprint analysis of Ophioglossum by HPLC. Chin J Exp Tradit Med Formulae. 22(06):61–65.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Lu W, Chen SW, Dong XT. 2009. Descriptions and microscopic identification of Ophioglossum vulgatum. J Fujian Univ Tradit Chin Med. 19(03):10–13.
- Thiers B. 2021. (continuously updated). Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/.
- Yang L, Li Y, Wei Y, Chen Z, Zhang F. 2019. Pharmacognosy research of Ophioglossum vulgatum. Northwest Pharm J. 34(05):569–573.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhang L, Fan XP, Petchsri S, Zhou L, Pollawatn R, Zhang X, Zhou XM, Ngan TL, Knapp R, Chantanaorrapint S, et al. 2020. Evolutionary relationships of the ancient fern lineage the adder's tongues (Ophioglossaceae) with description of Sahashia gen. nov. Cladistics. 36(4):380–393.
- Zhu T, Zeng BT, Wang TX, Zhou LJ, Yang HJ. 2014. Research progress on the rare medicinal plant Ophioglossum vulgatum. Anhui Agric Sci. 42(14):4226–4228.
