Abstract
The complete sequence of the mitochondrial genome of Balanus trigonus Darwin, 1854 was examined using next-generation sequencing analysis. The complete mitogenome of B. trigonus has 15,336 bp in length and comprises 37 genes, namely, 13 protein-coding genes (PCGs), 22 tRNAs, and two rRNAs. Both the gene order and characteristics are consistent with those of other species within the family Balanidae. Phylogenetic analysis based on complete mitogenomes revealed taxonomic relationships among members of the family Balanidae.
Barnacles of the genus Balanus Costa, 1778, within the family Balanidae in the order Balanomorpha, have a worldwide distribution that encompasses temperate and subtropical areas. To date, a total of 88 Balanus species have been described globally, of which 10 species have been reported from Korea (Kim Citation2011; Kim et al. Citation2020). Currently, however, the entire mitochondrial genome of only one (Balanus balanus) of these 10 species has been sequenced (Shen et al. Citation2014). Herein, we report the complete mitogenome of a further species, Balanus trigonus (MZ049958), which will contribute to assessments of the evolutionary relationships among barnacles.
Specimen of B. trigonus was collected in Tongyeong, Korea (34°49′41.8″N, 128°26′06.7″E) on 17 January 2021, at depths of between 0 and 1 m, and was subsequently identified based on morphological studies (Kim Citation2011; Kim et al. Citation2019). The voucher specimen (MABIK CR00248069) has been deposited in a deep freezer (−80 °C) of the National Marine Biodiversity Institute of Korea (Seongjun Bae, [email protected], Seocheon, Korea). Total genomic DNA was extracted from the specimen using a DNeasy Blood & Tissue DNA kit (Qiagen, Hilden, Germany), from which a genomic library was constructed using a QIAseq FX single-cell DNA library kit (Qiagen, Hilden, Germany) using paired-end reading. Next-generation sequencing analysis was conducted using an Illumina HiSeq 4000 system (Illumina Inc., USA). The complete mitogenome was reconstructed using Geneious Prime 2020.11.0 (Biomatters Ltd, Auckland, New Zealand).
The mitogenome of B. trigonus is 15,336 bp in length and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes. The overall nucleotide composition is 37.1% A, 15.6% C, 11.6% G, and 35.7% T. A majority of the PCG start codons are ATG (COX2, COX3, CYTB, ND2, ND5, and ND6), whereas the ATP6, ND1, and ND4 genes start with ATA, and ATT is the start codon for ATP8 and ND3. Exceptionally, AAA and GTG are used as alternative start codons for COX1 and ND4L, respectively. Notably, B. balanus (KM660676) similarly uses AAA as an alternative start codon for COX1. The most common stop codon is TAA (ATP6, ATP8, COX1, ND1, ND2, ND4L, ND5, and ND6), followed by TAG (CYTB). The remaining four PCGs (COX2, COX3, ND3, and ND4) were found to have an incomplete stop codon ‘T––’.
The dataset used for phylogenetic analysis included the 13 PCGs from 28 species in the 12 families and those from the three barnacles Lepas australis (NC_025295), Lepas anserifera (NC_026576) and Glyptelasma annandalei (MH891848) were used as outgroups. The best-fit substitution was estimated using jModelTest 2.1.1 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012). Maximum likelihood (ML) analysis was conducted using PhyML 3.1, based on the TVM + I + G model with 1,000 replications of bootstrap assembly (Guindon et al. Citation2010). In the ML tree thus obtained, B. trigonus was found to cluster as a sister group with Fistulonalanus albicostatus (MK617531) and Amphibalanus amphitrite (KF588709), which are related species within the family Balanidae.
Previously considered to be a single monophyletic, Archaeobalanidae and Balanidae have been combined into one large family in a recent phylogeny study through molecular evidence (Tsang et al. Citation2017; Chan et al. Citation2021). In our results, Armatobalanus alliusm (presently in Balanidae) and Pyrgomatidae form a monophyletic clade (). This pattern is consistent with those reported previously (Chan et al. Citation2021; Ji et al. Citation2021; Mao et al. Citation2021). However, Simon-Blecher et al. Citation2007 and Tsang et al. Citation2014 stated A. allium should be pyrgomatid barnacles based on morphology and molecular evidence. A revision of the taxonomic status of A. allium will be a further research direction.
Figure 1. Phylogenetic tree inferred by maximum-likelihood using of 13 protein-coding genes of 28 barnacles mitochondrial genomes, including B. trigonus (MZ049958). Bootstrap support values based on 1,000 replicates are displayed on each node as >70.
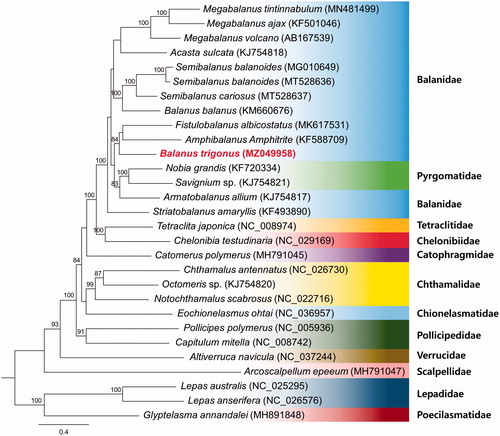
We found the molecular phylogenetic relationships between Balanidae and Pyrgomatidae to be inconsistent with the current classification based on morphological features, thereby highlighting the necessity for further clarification of the phylogenetic classification between Balanidae and Pyrgomatidae, and the need for further research regarding morphological reclassification. In this regard, the mitogenome sequence obtained in the present study will serve as valuable a genomic resource, contributing to further molecular studies on the evolution of the family Balanidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ049958. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA717899, SRR14270317, and SAMN18515342, respectively.
Additional information
Funding
References
- Chan BKK, Dreyer N, Gale AS, Glenner H, Ewers-Saucedo C, Pérez-Losada M, Kolbasov GA, Crandall KA, Høeg JT. 2021. The evolutionary diversity of barnacles, with an updated classification of fossil and living forms. Zoological J Linnean Soc. 160.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Ji N, Ge T, Mao S, Zhang M, Mao N, Cai Y, Shen X. 2021. The first mitochondrial genome of Tetraclita kuroshioensis (Crustacea: Sessilia) from China: insight into the phylogeny within Cirripedia. Mitochondrial DNA B Resour. 6(3):988–989.
- Kim I-H. 2011. Invertebrate fauna of Korea, barnacles (Arthropoda: Crustacea: Cirripedia). National Institute of Biological Resources, Ministry of Environment. 21(6):1–144.
- Kim HK, Chan BKK, Lee S, Kim W. 2020. Biogeography of intertidal and subtidal native and invasive barnacles in Korea in relation to oceanographic current ecoregions and global climatic changes. J Mar Biol Ass. 100(7):1079–1091.
- Kim H, Lee S, Bs M, Kim W. 2019. Report on the current status of the distribution of invasive barnacles in marine national park areas of Korea. J National Park Res. 10(2):249–257.
- Mao S, Ge T, Cai Y, Ji N, Kong X, Shen X. 2021. The mitochondrial genome of Chthamalus malayensis (Sessilia: Chthamalidae) and its molecular phylogeny within Cirripedia. Mitochondrial DNA B Resour. 6(2):643–644.
- Shen X, Tsoi KH, Cheang CC. 2014. The model barnacle Balanus balanus Linnaeus, 1758 (Crustacea: Maxillopoda: Sessilia) mitochondrial genome and gene rearrangements within the family Balanidae. Mitochondrial DNA Part A. 27(3):2112–2114.
- Simon-Blecher N, Huchon D, Achituv Y. 2007. Phylogeny of coral-inhabiting barnacles (Cirripedia; Thoracica; Pyrgomatidae) based on 12S, 16S and 18S rDNA analysis. Mol Phylogenet Evol. 44(3):1333–1341.
- Tsang LM, Chu KH, Nozawa Y, Chan BKK. 2014. Morphological and host specificity evolution in coral symbiont barnacles (Balanomorpha: Pyrgomatidae) inferred from a multi-locus phylogeny. Mol Phylogenet Evol. 77:11–22.
- Tsang LM, Shen X, Cheang CC, Chu KH, Chan BKK. 2017. Gene rearrangement and sequence analysis of mitogenomes suggest polyphyly of Archaeobalanid and Balanid barnacles (Cirripedia: Balanomorpha). Zool Scr. 46(6):729–739.
