Abstract
The mitochondrial genome (mitogenome) of Saussurella borneensis (Orthoptera: Tetrigoidea) was determined and analyzed. The complete mitochondrial genome is 16,006 bp in length, consisting of 37 genes, including 13 protein-coding (PCGs), 22 tRNA, 2 rRNA genes as well as an A + T-rich region. Ten PCGs initiated with a typical ATN codon (one with ATC, two with ATA, two with ATT, and five with ATG) and 13 terminated with complete stop codons. The overall nucleotide composition was 42.97% for A, 17.61% for C, 11.62% for G, and 27.8% for T. Phylogenetic analysis of S. borneensis fully resolved it in a basal branch sister to Tripetaloceroides tonkinensis. This data increase the bioinformatics of the Tetrigidae, and improves our understanding of the phylogenetic status of S. borneensis in the Tetrigoidea.
Saussurella borneensis Hancock, 1912, is classified in the subfamily Batrachidinae of Orthoptera. This subfamily currently contains 3 tribes and 25 known genera worldwide; the genus Saussurella Bolivar includes twelve species endemic to Southeast Asia (Deng Citation2016; Zha et al. Citation2020). However, there is little information on the systematic position of S. borneensis within the Tetrigidae (Zhang et al. Citation2020). To date, no mitochondrial sequence has been reported for the Batrachidinae (NCBI, last visited on May 2021). To further advance the evolutionary studies of the Batrachidinae, we sequenced, assembled and performed a phylogenetic analysis of the mitochondrial genome of S. borneensis (GenBank accession No. MZ169555) to contribute to the evolutionary systematics of the Batrachidinae.
The samples of S. borneensis were collected from Nonggang Nature Reserve in Guangxi province of China (22.474261°N, 106.957389°E) in May 2020 and the voucher specimen was deposited in Entomological Museum of Hechi University (EMHU) (the voucher No. bb1, Hechi University, Yizhou, China, Deng WA, [email protected]). Total genomic DNA was extracted from the legs of an adult specimen of S. borneensis using the DNeasy Blood & Tissue Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. The genomic DNA was sequenced using 150 bp PE on the Illumina Novaseq platform (Personalbio, Shanghai, China). The mitogenome was de novo assembled using A5-miseq v20150522 (Coil et al. Citation2015) and SPAdes version 3.9.0 (Bankevich et al. Citation2012), and all genes were annotated with the MITOS Web Server (Bernt et al. Citation2013).
The complete mitogenome of S. borneensis is 16,006 bp, and contains 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNA (rrnS and rrnL), and an A + T-rich region. The composition of the mitogenome is 42.97% A, 17.61% C, 11.62% G, and 27.8% T, showing an A + T bias (70.77%). Nine PCGs and fourteen tRNAs were transcribed from the majority strand, while the remaining four PCGs (ND1, ND4, ND4L, and ND5), eight tRNAs and two rRNAs were located on the minority strand. Ten PCGs initiated with a typical ATN codon (one with ATC, two with ATA, two with ATT and five with ATG), whereas the ND1, ND4, and ND4L genes started with TTG. Thirteen terminated with complete stop codons (one with AAC, AAG, ACA, ATG, ATT, GCA, GTA, TGA, and TGT, respectively, two with GAA and two with TTA). Twenty-two tRNA genes were found interspersed in the mitogenomes of S. borneensis, which ranged in size from 62 bp (trnH) to 70 bp (trnK and trnV). The two rRNA genes, i.e. rrnS (742 bp) and rrnL (1371 bp), were located between the trnL1 and an A + T-rich region, and separated by the trnV gene. The gene composition, order and orientation of S. borneensis was the same as the mitogenomes of other tetrigid species, such as Tripetaloceroides tonkinensis (MW770353) and Tetrix japonica (JQ340002).
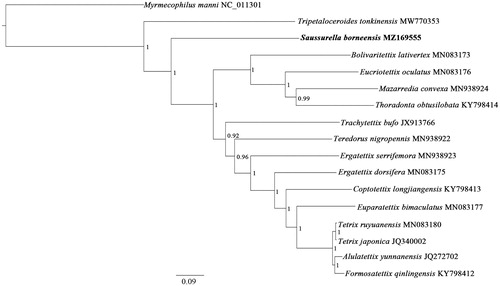
To validate the phylogenetic position of S. borneensis, a Bayesian Inference (BI) tree was constructed () using MrBayes version 3.2.6 (Ronquist et al. Citation2012) with 13 PCGs (10,734 bp) from the mitogenomes of sixteen tetrigid species and one outgroup taxon (Myrmecophilus manni), respectively. As shown in , T. tonkinensis occupied the most basal position, followed by the second branch, S. borneensis, which was positioned as a sister group to the remaining Tetrigoidea (posterior probability, PP = 1). These data, suggesting that T. tonkinensis is the earliest species within Tetrigoidea, followed by S. borneensis, which was consistent with the results of the molecular phylogenetic analyses conducted by earlier study showing that S. borneensis is an earlier species within Tetrigoidea (Zhang et al. Citation2020). Moreover, sixteen tetrigid species were grouped in a clade with strong support (PP = 1), which suggested Tetrigidae is monophyletic.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MZ169555. The associated BioProject, SRA and BioSample numbers are PRJNA743276, SRR15031231, and SAMN20014183, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Deng WA. 2016. Taxonomic study of Tetrigoidea from China [Ph.D. dissertation]. Wuhan, China: Huazhong Agricultural University.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Zha LS, Chomnunti P, Ding JH, Zhang HJ. 2020. Saussurella brevifrons sp. nov. from northern Thailand, with taxonomic notes for the genus (Orthoptera: Tetrigidae). Entomol News. 129(4):367–376.
- Zhang RJ, Zhao CL, Wu FP, Deng WA. 2020. Molecular data provide new insights into the phylogeny of Cladonotinae (Orthoptera: Tetrigoidea) from China with the description of a new genus and species. Zootaxa. 4809(3):547–559.