Abstract
Castanopsis sieboldii (Makino) Hatus is an evergreen tree that distributes in Eastern Asia including Islands of Korea and Japan. The chloroplast genome of C. sieboldii was successfully sequenced. Its length is 160,705 bp long (GC ratio is 36.8%) and has four subregions: 90,821 bp of large single copy (34.6%) and 19,014 bp of small single copy (30.8%) regions are separated by 25,075 bp of inverted repeat (42.8%) regions including 134 genes (89 protein-coding genes, eight rRNAs, and 37 tRNAs). Interspecific variations of Castanopsis are at a moderate level in comparison to those of the other genera. Phylogenetic trees show that C. sieboldii chloroplast genome was clustered with the other two Castanopsis species.
Castanopsis sieboldii (Makino) Hatus, belonging to Fagaceae, is a species of an evergreen tree that distributes in subtropical Eastern Asia, including Islands in Western and Southern Korea (Park, An, et al. Citation2020) and Honshu, Shikoku, and Kyushu Islands in Japan (Yamazaki Citation1987). This species is considered as a climax species in an evergreen forest (Yamanaka Citation1966). The genetic diversity of C. sieboldii in Japan is relatively high (Aoki et al. Citation2014): one of the major reasons is that they have been isolated in islands. It is one of hot topics to understand the origin of the island species based on chloroplast genomes, such as founder effects (Del Valle et al. Citation2019), multiple introductions in the islands (Nock et al. Citation2019), or the origin of endemic species in the islands (Yang et al. Citation2018; Park, Bae, et al. Citation2021). To understand its genetic diversity of C. sieboldii in Korea, as the first step, we completed the first chloroplast genome of C. sibboldii from the sample isolated in one of the islands in Korea, Oenalodo Island.
Total DNA of C. sieboldii collected at Singeum-gil, Bongnae-myeon, Goheung-gun, Jeollanam-do, Republic of Korea (34°28'18.58′'N, 127°28'04.41′'E) was extracted from fresh fruits with a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The voucher was deposited in the InfoBoss Cyber Herbarium (IN; http://herbarium.infoboss.co.kr/; Voucher number: IB-01087; Contact: Suhyeon Park; [email protected]). Genome sequencing was conducted using NovaSeq6000 at Macrogen Inc., Korea, and de novo assembly and sequence confirmation were done by Velvet v1.2.10 (Zerbino and Birney Citation2008), GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) in the Genome Information System (GeIS; http://geis.infoboss.co.kr/) which has been utilized in the previous organelle genomic studies (Park, Kim, Xi, Nho, et al. Citation2019; Park, Park, Kim, et al. Citation2019; Park, Yun, Oh, et al. Citation2019; Park, Lee, et al. Citation2021; Park, Min, et al. Citation2021; Joo et al. Citation2020; Park and Oh Citation2020; Kim et al. Citation2021). Geneious Prime® v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Castanopsis fargesii chloroplast (NC_047230; Ye et al. Citation2019).
The chloroplast genome of C. sieboldii (MZ028444) is 160,705 bp (GC ratio is 36.8%) and has four subregions: 90,821 bp of large single copy (34.6%) and 19,014 bp of small single copy (SSC; 30.8%) regions are separated by 25,705 bp of the inverted repeat (IR; 42.8%). It contains 134 genes (89 protein-coding genes, eight rRNAs, and 37 tRNAs); 20 genes (nine protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in the IR regions.
Based on pair-wise alignments against chloroplast genomes of C. fargesii and C. concinna, distributed in Southern China (Sun et al. Citation2014; Daniel Hinsinger and Sergej Strijk Citation2017), 520 single nucleotide polymorphisms (SNPs) and 144 insertion and deletion (INDEL) regions covering 709 bp and 220 SNPs and 125 INDEL regions covering 533 bp were identified, respectively. Numbers of these interspecific variations are fewer in number than those of the four species of Viburnum, displaying 944 to 1295 SNPs and 1697-bp to 3080-bp INDELs (Park, Xi, et al. Citation2020), and Potentilla micrantha and Potentilla centigrana (1570 SNPs and 3,451-bp INDELs; Ferrarini et al. Citation2013; Park et al. Citation2019), and Potentilla freyniana and Potentilla chinensis (1236 SNPs and 2295-bp INDELs; Park et al. Citation2019; Dang et al. Citation2020). Moreover, they are similar or higher than the numbers of intraspecific variations identified from the samples between Korea and China (Heo et al. Citation2019; Oh et al. Citation2019a, Citation2019b; Park, Kim, Xi, Oh, et al. Citation2019; Park, Suh, et al. Citation2020; Heo et al. Citation2020; Oh and Park Citation2020). These results indicate that the numbers of interspecific variations identified from the three Castanopsis species are at a moderate level.
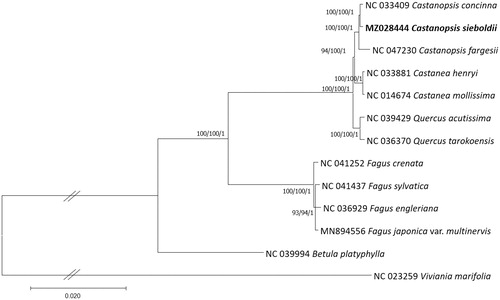
Thirteen Fagaceae chloroplast genomes including one outgroup species, Betula platyphylla, were used for constructing bootstrapped Maximum-Likelihood (ML), Neighbor-joining (NJ), and Bayesian Inference (BI) phylogenic trees using MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.6 (Ronquist et al. Citation2012), respectively, after aligning whole chloroplast genomes by MAFFT v7.450 (Katoh and Standley Citation2013). A heuristic search was used with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites to construct ML and NJ phylogenetic trees with default values for other options using MEGA X. Bootstrap analyses with 1000 and 10,000 pseudoreplicates were conducted for ML and NJ trees, respectively. The GTR model with gamma rates was used as a molecular model and Markov-chain Monte Carlo algorithm was employed for 1,100,000 generations, sampling trees every 200 generations, with four chains running simultaneously for BI tree. Three phylogenetic trees display that three Castanopsis species are clustered in one clade and are congruent to each other with high supportive values of ML, NJ, and BI (). In addition, the topology of Castanopsis, Castanea, Quercus, and Fagus genera in the phylogenetic tree is congruent to the previous phylogenetic and morphological study (Manos et al. Citation2008). Taken together, our chloroplast genome is useful to investigate phylogenetic relationships of C. sieboldii as well as its genetic diversities along with geographical distribution.
Figure 1. Maximum-Likelihood (bootstrap repeat is 1000), Neighbor-joining (bootstrap repeat is 10,000), and Bayesian inference (Number of generations is 1,100,000) phylogenetic trees of Fagaceae thirteen chloroplast genomes. Phylogenetic tree was drawn based on Maximum-Likelihood tree. The numbers above branches indicate bootstrap support values of Maximum-Likelihood, Neighbor-joining, and Bayesian inference phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
Chloroplast genome sequence can be accessed via accession number of MZ028444 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA724746, SAMN18857600, and SRR14316975, respectively.
Additional information
Funding
References
- Aoki K, Ueno S, Kamijo T, Setoguchi H, Murakami N, Kato M, Tsumura Y. 2014. Genetic differentiation and genetic diversity of Castanopsis (Fagaceae), the dominant tree species in Japanese broadleaved evergreen forests, revealed by analysis of EST-associated microsatellites. PloS One. 9(1):e87429.
- Dang H, Xu J, Jin Z. 2020. The complete chloroplast genome of Potentilla chinensis. Mitochondrial DNA Part B. 5(2):1180–1181.
- Daniel Hinsinger D, Sergej Strijk J. 2017. Complete chloroplast genome sequence of Castanopsis concinna (Fagaceae), a threatened species from Hong Kong and South-Eastern China. Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):65–66.
- Del Valle JC, Casimiro-Soriguer I, Buide M, Narbona E, Whittall JB. 2019. Whole plastome sequencing within Silene section Psammophilae reveals mainland hybridization and divergence with the Balearic Island populations. Front Plant Sci. 10:1466.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, et al. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics. 14(1):612–670.
- Heo K-I, Kim Y, Maki M, Park J. 2019. The complete chloroplast genome of mock strawberry, Duchesnea indica (Andrews) Th. Wolf (Rosoideae). Mitochondrial DNA Part B. 4(1):560–562.
- Heo K-I, Park J, Xi H, Min J. 2020. The complete chloroplast genome of Agrimonia pilosa Ledeb. isolated in Korea (Rosaceae): investigation of intraspecific variations on its chloroplast genomes. Mitochondrial DNA B Resour. 5(3):2264–2266.
- Joo S, Lee J, Lee D-Y, Xi H, Park J. 2020. The complete mitochondrial genome of the millipede Epanerchodus koreanus Verhoeff, 1937 collected in limestone cave of Korea (Polydesmidae: Polydesmida). Mitochondrial DNA B Resour. 5(4):3845–3847.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim M, Xi H, Park J. 2021. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: intraspecific characteristics of GATA transcription factors. PLoS One. 16(5):e0252181.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Manos PS, Cannon CH, Oh S-H. 2008. Phylogenetic relationships and taxonomic status of the paleoendemic Fagaceae of western North America: recognition of a new genus. Notholithocarpus. Madroño. 55(3):181–190.
- Nock CJ, Hardner CM, Montenegro JD, Ahmad Termizi AA, Hayashi S, Playford J, Edwards D, Batley J. 2019. Wild origins of macadamia domestication identified through intraspecific chloroplast genome sequencing. Front Plant Sci. 10:334.
- Oh S-H, Park J. 2020. The complete chloroplast genome of Euscaphis japonica (Thunb.) Kanitz (Staphyleaceae) isolated in Korea. Mitochondrial DNA B Resour. 5(3):3753–3769.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019b. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA Part B. 4(1):626–627.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019a. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondrial DNA B Resour. 4(2):2692–2693.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Bae Y, Kim B-Y, Nam G-H, Park J-M, Lee BY, Suh H-J, Oh S-H. 2021. The complete chloroplast genome of Campanula takesimana Nakai from Dokdo Island in Korea (Campanulaceae). Mitochondrial DNA B Resour. 6(1):135–137.
- Park J, Heo K-I, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla centigrana Maxim. (Rosaceae). Mitochondrial DNA Part B. 4(1):688–689.
- Park J, Heo K-I, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla freyniana Bornm. (Rosaceae). Mitochondrial DNA B Resour. 4(2):2420–2421.
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. 2019. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4(1):1541–1542.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4(1):1038–1040.
- Park J, Lee J, Lee W. 2021. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Leonurus japonicus in Korea. Mitochondrial DNA B Resour. 6(1):62–65.
- Park J, Min J, Kim Y, Chung Y. 2021. The comparative analyses of six complete chloroplast genomes of morphologically diverse Chenopodium album L. (Amaranthaceae) collected in Korea. Int J Genomics. 2021:6643444.
- Park J, Oh S-H. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): intraspecific variations on chloroplast genome. Mitochondrial DNA Part B. 5(2):1868–1869.
- Park J, Park J, Kim J-H, Cho JR, Kim Y, Seo BY. 2019. The complete mitochondrial genome of Micromus angulatus (Stephens, 1836)(Neuroptera: Hemerobiidae). Mitochondrial DNA Part B. 4(1):1467–1469.
- Park J, Suh Y, Kim S. 2020. A complete chloroplast genome sequence of Gastrodia elata (Orchidaceae) represents high sequence variation in the species. Mitochondrial DNA B Resour. 5(1):517–519.
- Park J, Xi H, Oh S-h. 2020. Comparative chloroplast genomics and phylogenetic analysis of the Viburnum dilatatum complex (Adoxaceae) in Korea. Korean J Pl Taxon. 50(1):8–16.
- Park J, Yun N, Oh S-H. 2019. The complete chloroplast genome of an endangered species in Korea, Halenia corniculata (L.) Cornaz (Gentianaceae). Mitochondrial DNA Part B. 4(1):1539–1540.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sun Y, Hu H, Huang H, Vargas-Mendoza CF. 2014. Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genet Genomes. 10(6):1531–1539.
- Yamanaka T. 1966. Problem of Castanopsis cuspidata Schottky. Bull Fac Educ Kochi Univ. 18:65–73.
- Yamazaki H. 1987. A taxonomical revision of Castanopsis cuspidata (Thunb) Schottky and the allies in Japan and Taiwan 1. J Jpn Bot. 62:289–298.
- Yang JY, Pak J-H, Kim S-C. 2018. The complete plastome sequence of Rubus takesimensis endemic to Ulleung Island, Korea: insights into molecular evolution of anagenetically derived species in Rubus (Rosaceae). Gene. 668:221–228.
- Ye X-M, Guo Y-P, Lei X-G, Sun R-X. 2019. The complete chloroplast genome of Castanopsis fargesii Franch.(Fagaceae). Mitochondrial DNA Part B. 4(1):1656–1657.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12 Suppl 14(14):S2.
