Abstract
Rotheca myricoides (Hochst.) Steane & Mabb. is a plant species used in traditional medicine for the management of diabetes in the lower eastern part of Kenya (Kitui, Machakos and Makueni Counties, Kenya) that is mainly inhabited by the Kamba community. The complete chloroplast genome sequence of R. myricoides was assembled from the whole genome Illumina sequencing data. The genome was 150,596 bp in length, contained an SSC region of 17,237 bp and LSC region of 83,489 bp, separated by IRs of 24,935 bp, each. The genome contained 114 unique genes, including 80 PCGs, 4 rRNA genes, and 30 tRNA genes. In addition, 18 genes contained one or two introns, including 10 PCG genes with a single intron, 2 PCG genes harboring two introns, and 6 tRNA genes harboring a single intron. Phylogenetic analysis supported R. myricoides had the closest genetic relationship with Rotheca serrata and clustered with the Rotheca family species.
R. myricoides is used globally as a traditional medicinal plant species belonging to the genus Rotheca the largest genus of the family Labiatae (Bashwira and Hootele Citation1988). It is used for management of diabetes in the lower eastern part of Kenya. A leaf decoction is prepared by boiling and a cup (250 ml) is taken daily (Keter and Mutiso Citation2012). Other ethnobotanical uses of this species include epilepsy, arthritis, typhoid, cough, eye problems, tonsillitis, rheumatism, gonorrhea (Moshi et al. Citation2012), cancer (Esubalew et al. Citation2017), malaria (Mukungu et al. Citation2016), dysmenorrhea, sterility, impotence, cough, furunculosis, inflammation, and snakebites (Richard et al. Citation2011). It is traditionally brewed and drank as a tea product in Asian countries to relieve swelling and pain. In China, R. myricoides is mainly distributed in Guangdong provience. In this study, the complete plastid genome sequence of R. myricoides is a useful resource and provides additional valuable data for the phylogenetic study of Rotheca in the future.
The fresh leaves of R. myricoides were collected in Zhanjiang (110.3 E, 21.2 N; Guangdong, China), and the voucher specimen (RM772543) deposited at Pharmaceutical Laboratory in Xi'an International University (Siyi Zhang, [email protected]). The total genomic DNA was extracted with modified CTAB protocol. Subsequently, the integrity and quality of the DNA werechecked through agarose gel electrophoresis and spectrophotometry using a Nanodrop 2000 instru-ment (Thermo Scientific, DE, USA), respectively. Then the genomic DNA of R. myricoides was used for the subsequent shotgun library construction and High-throughput sequencing on the Illumina HiSeq X Ten Sequencing System (Illumina, CA, USA).
The data were assembled R. myricoides chloroplast genome sequence with length 150, 596 bp. The cp genome was showed a typical quadripartite structure, consisted of a large single copy region with 83,489 bp (LSC) and small single copy region with 17,237 bp (SSC), and separated by a pair of inverted repeatregions with 24,935 bp (IRs), each. The whole genome sequence GC content is 38.6%, while in LSC, SSC and IRs (IRA + IRB) are 36.37%, 32.8% and 43.7%, respectively. A total of 113 genes were predicted in the whole chloroplast genome of R. myricoides, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. In addition, 18 genes contain one or two introns, which include 10 PCG genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16) possessing a single intron, 2 PCG genes (clpP, ycf3) harboring two introns, and 6 tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) harboring a single intron.
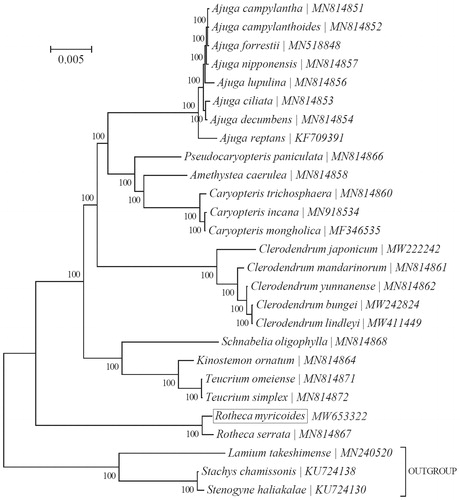
To ascertain its phylogenetic placement within the family Rotheca, the Bayesian Inference (BI) phylogenetic tree was reconstructed with the MRBayes V3.1.2 (Ronquist and Huelsenbeck Citation2003) integrated with Topali V2.5 (Milne et al. Citation2009) based on 15 genes (atpA, atpB, matK, ndhA, ndhH, psaB, psbA, psbB, psbC, psbD, rbcL, rpoA, rpoB, rpoC1 & rpoC2) in the complete chloroplast genome of R. myricoides and 26 other species. As expected, R. myricoide was close to R. serrata formed a strongly monophyletic clade (). Our findings suggest that the high-level taxonomy of the family Rotheca may need further investigations.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the US National Center for Biotechnology Information (NCBI database) at https://www.ncbi.nlm.nih.gov/, reference number: MW653322, and the associated BioProject, SRA, and Bio-Sample numbers are PRJNA724817, SRR14322715 and SAMN18858889, respectively.
Additional information
Funding
References
- Bashwira S, Hootele C. 1988. Myricoidine and dihydromyricoidine, two new macrocyclic spermidine alkaloids from Clerodendrum myricoides. Tetrahedron. 44(14):4521–4526.
- Esubalew ST, Belet A, Lulekal E, et al. 2017. Review of ethnobotanical and ethnopharmacological evidences of some ethiopian medicinal plants traditionally used for the treatment of cancer. Ethiopian J Health Develop. 31(3):161–187.
- Keter L, Mutiso P. 2012. Ethnobotanical studies of medicinal plants used by Traditional Health Practitioners in the management of diabetes in Lower Eastern Province, Kenya. J Ethnopharmacol. 139(1):74–80.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Moshi MJ, Otieno DF, Weisheit A. 2012. Ethnomedicine of the Kagera region, north western Tanzania. Part 3: plants used in traditional medicine in Kikuku village, Muleba district. J Ethnobiol Ethnomed. 8:14.
- Mukungu N, Abuga K, Okalebo F, Ingwela R, Mwangi J. 2016. Medicinal plants used for management of malaria among the Luhya community of Kakamega East sub-County, Kenya. J Ethnopharmacol. 194:98–107.
- Richard S, Maciuk A, Banzouzi J, Champy P, Figadere B, Guissou IP, Nacoulma OG. 2011. Mutagenic effect, antioxidant and anticancer activities of six medicinal plants from Burkina Faso. Natural Prod Res. 26(6):575–579.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.