Abstract
In this study, we report the complete mitochondrial genome of Stichopus chloronotus. The mitogenome was 16,247 base pairs (58.55% A + T content) in length, comprising a total of 37 genes, including 13 protein-coding genes, 22 transfer RNA genes and 2 ribosomal RNA genes. To resolve the phylogenetic position of S. chloronotus, we analyzed all mitochondrial protein-coding genes from 27 species within the Echinodermata. The results showed that S. chloronotus belonged to the family Stichopodidae and was more closely related to tropical Stichopus species (S. horrens and S. monotuberculatus) than to other species. Our results will be useful for evolutionary analysis of sea cucumber species.
Sea cucumbers are an important group of benthic organisms comprising abundant and diverse ecologically and economically important species (Liao Citation1997). These organisms are widely distributed in oceans worldwide from depths of ten thousand meters in the sea or trenches to intertidal zones. They mainly live-in reefs, sediment and seaweed and are often overgrown (Gallo et al. Citation2015; Mu et al. Citation2018; Yang et al. Citation2019). Stichopus chloronotus (S. chloronotus) is widely distributed in the Indo-Western Pacific region, and mainly inhabits in the Xisha Islands of Hainan Province in China (Liao Citation1997). S. chloronotus contains sticholoroside, stichostatin I, fucosylated chondroitin sulfate (Mou et al. Citation2020), fatty acids (Fredalina et al. Citation1999) and other compounds. The active ingredients of S. chloronotus were reported to expedite the wounds healing (Mohamed et al. Citation2015). Complete mitochondrial sequencing is an important method for analyzing phylogenetic relationships and identifying species of sea cucumbers (Rowe and Richmond Citation2004; Cheng et al. Citation2019, Citation2020). However, the mitochondrial genome of S. chloronotus has not yet been revealed. In this study, we constructed a phylogenetic tree of 27 species in the sea cucumber family. These molecular data will be useful for the evolutionary analysis of tropical sea cucumbers.
The specimen was obtained from Sanya City, Hainan Province, China (N18°2021′, E109°4708′). Total DNA was extracted from longitudinal muscle tissue by using a TIANamp Marine Animals DNA Kit (Tiangen Biochemistry Technology Co., Ltd., China) and deposited at Guangdong Academy of Agricultural Sciences in Guangzhou, China (http://www.gdias.net/, Xiaoying Chen is the contact person [email protected]) under the voucher number GDAAS-IAS-AQUA-CA-202008102. The DNA was sequenced on an Illumina HiSeq 4000 sequencing platform (150-bp paired-end reads were generated) at Shenzhen Huitong Biotechnology Co., Ltd., China (Li et al. Citation2021). Raw sequence reads were edited using the NGS QC Tool Kit v2.3.3 and assembled into contigs using the de novo assembler SPAdes 3.11.0 (Dmitry et al. Citation2016), and the complete mitogenome was deposited on the NCBI website (https://www.ncbi.nlm.nih.gov/genbank/) with the accession number MW218897.1. The protein-coding genes (PCGs; tRNA and rRNA genes) were annotated by using MITOS Web Server BETA (http://bloodymary.bioinf.uni-leipzig.de/mitos/index.py). The tRNA genes were identified by using RNAscan-SE software (Lowe and Chan Citation2016). The phylogenetic relationships were reconstructed with the PCGs by means of maximum-likelihood (ML) (GTR + G + I model) analysis by using MEGA-X software with 1000 replicates (Kumar et al. Citation2018; Li et al. Citation2021).
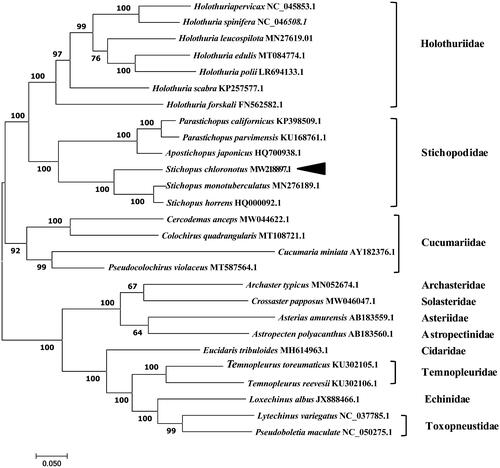
The complete mitogenome of S. chloronotus was 16,247 bp in length (GenBank accession number: MW218897.1) and contained the typical set of 13 PCGs, 22 tRNA genes, 2 rRNA genes, and one putative control region. The overall base composition of the mitogenome was estimated to be 31.48% A, 27.07% T, 25.39% C and 16.06% G, with a high A + T content of 58.55%. In the phylogenetic tree, S. chloronotus was distinct from the other species, forming an independent branch (). The results showed that S. chloronotus belonged to the family Stichopodidae, and we revealed for the first time that S. chloronotus was more closely related to the tropical Stichopus species S. horrens and S. monotuberculatus than to other echinoderm species.
Disclosure statement
No potential conflicts of interest were reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of the NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. MW218897.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA704715, SRR13787362and SAMN18043597, respectively.
Additional information
Funding
References
- Cheng CH, Wu FF, Ren CH, Jiang X, Wu XF, Huang W, Hu CQ. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of a tropical sea cucumber, Holothuria fuscocinerea. Mitochondrial DNA B Resour. 5(3):2677–2678.
- Cheng CH, Yu ZH, Ren CH, Jiang X, Zhang X, Wu XF, Huang W, Hu CQ. 2019. The complete mitochondrial genome of a tropical sea cucumber, Holothuria leucospilota. Mitochondrial DNA B Resour. 4(2):3118–3119.
- Dmitry A, Anton K, Jeffrey SM, Pavel AP. 2016. HYBRIDSPADES: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32:7.
- Fredalina BD, Ridzwan BH, Abidin AA, Kaswandi MA, Zaiton H, Zali I, Kittakoop P, Jais AM. 1999. Fatty acid compositions in local sea cucumber, Stichopus chloronotus, for wound healing. Gen Pharmacol. 33(4):337–340.
- Gallo ND, Cameron J, Hardy K, Fryer P, Bartlett DH, Levin LA. 2015. Submersible- and lander-observed community patterns in the Mariana and New Britain trenches: influence of productivity and depth on epibenthic and scavenging communities. Deep Sea Res Part I Oceanogr Res Pap. 99:119–133.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H, Liu J, Wang S, Huang W. 2021. The complete mitochondrial genome of Pink warty sea cucumber (Cercodemas anceps Selenka, 1867). Mitochondrial DNA B Resour. 6(3):959–961.
- Liao Y. 1997. Fauna Sincia: phylum Echinodermata class Holothuroidea. Beijing: Science Press.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mohamed IN, Mazlan M, Shuid AN. 2015. Sea cucumber (Stichopus chloronotus) to expedite healing of minor wounds. World Acad Sci Eng Technol Int J Biomed Biol Eng. 9(11):1.
- Mou JJ, Li Q, Shi WW, Qi XH, Song WJ, Yang J. 2020. Chain conformation, physicochemical properties of fucosylated chondroitin sulfate from sea cucumber Stichopus chloronotus and its in vitro fermentation by human gut microbiota. Carbohydr Polym. 228:115310–115359.
- Mu WS, Liu J, Zhang HB. 2018. Complete mitochondrial genome of Benthodytes marianensis (Holothuroidea: Elasipodida: Psychropotidae): insight into deep sea adaptation in the sea cucumber. PLOS One. 13(11):e0208051.
- Rowe FWE, Richmond MD. 2004. A preliminary account of the shallowwater echinoderms of Rodrigues, Mauritius, western Indian Ocean. J of Natural Hist. 38(23):3273–3314.
- Yang QH, Lin Q, Yang FY, Wu JS, Lu Z, Li SK, Zhou C. 2019. Characterization of the complete mitochondrial genome of a holothurians species: Holothuria hilla (Holothuroidea: Holothuriidae). Mitochondrial DNA B Resour. 4(2):2847–2848.