Abstract
The chloroplast genome and evolutionary relationship analysis of Tulipa patens could provide fundamental genetic reference for its molecular breeding and biological research. The complete chloroplast genome of T. patens was sequenced and reported here. The genome was 152,050 bp in length, containing a pair of inverted repeated regions (26,330 bp) which were separated by a large single copy region of 82,184 bp, and a small single copy region of 17,206 bp. A total of 133 functional genes were annotated, including 87 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The phylogenetic relationships of 10 species indicated that T. patens was closely related to Tulipa sylvestris.
Keywords:
The Liliaceae family includes approximately 250 genera and 3500 species distributed worldwide. Tulips (genus Tulipa and family Liliaceae) are among the world's most well-known, beloved, and economically important ornamental plants (Li et al. Citation2017) and a variety of cultivars are grown for cut flowers, flowering potted plants, and landscaping (Van Tuyl and van Creij Citation2007). There are about 150 species of Tulipa, mainly distributed in Asia, Europe and North Africa, among which the Mediterranean to Central Asia is the most abundant (Xing et al. Citation2017), constituting a great genetic resource for the continued breeding and improvement of tulip cultivars (Xing et al. Citation2020). Chloroplasts are organelles that are essential for photosynthesis (Mehmood et al. Citation2020). The structure and composition of plastid genomes have become widely utilized in identifying unique genetic changes and evolutionary relationships of various groups of plants (Mehmood et al. Citation2020). Here, we sequenced the complete chloroplast genome of Tulipa patens (Genbank accession number: MT327740), to provide more primary genetic information for the phylogenetic relationship analysis of the genus and other relevant research.
In this study, T. patens were collected from Altay Region, Buerjing County, Xinjiang Province, China (48°41′48″ N, 87°02′03″ E) in September 2018. A specimen was deposited at Key Laboratory of Landscape Plants of Qinghai Province, Qinghai University, Xining, China (Xiu-Ting, JU, [email protected]) under the voucher number JXT-2019-YJX018. The total genomic DNA was extracted using leaves from the same plant. After DNA extraction, a library with an insertion size of 350 bp was constructed, and genomic sequencing was performed on the Illumina HiSeq Platform (Illumina, San Diego, CA) with a read length of 150 bp. The software SPAdes v.3.10.1 (Bankevich, et al. Citation2012) was employed to assemble the chloroplast genome, kmer uses 55, 87,121. Then, Prodigalv2.6.3 (https://www.github.com/hyattpd/Prodigal) was used to annotate the coding sequences (CDs) under the default parameter settings. Hmmer v3.1b2 (http://www.hmmer.org/) and Aragorn v1.2.38 (http://130.235.244.92/ARAGORN/) were respectively used to predict transfer RNA (tRNA) genes and ribosomal RNA (rRNA) genes under the default parameter settings.
The complete chloroplast genome of T. patens was 152,050 bp in length with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions (26,330 bp) that were separated by a large single copy (LSC) region of 82,184 bp, and a small single copy (SSC) region of 17,206 bp. The average coverage depth of the assembly was 1876.2875. The GC content of the whole complete chloroplast genome was 36.65%. A total of 133 functional genes were annotated, including 87 protein-coding genes (PCGs), 38 tRNA genes, and eight rRNA genes.
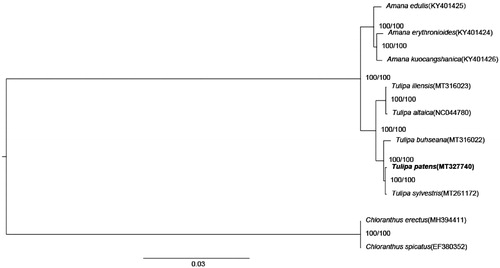
The complete chloroplast genomes of eight species from the Liliaceae family and two outgroups from the Chloranthaceae family were used to construct the maximum likelihood tree with 1000 bootstrap repeats (model: TVM + F+G4) by W-IQ-TREE (Trifinopoulos et al. Citation2016) after aligned by MAFFT 7 (Katoh and Standley Citation2013). The phylogenetic tree showed that T. patens was closely related to Tulipa sylvestris ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT327740.The associated BioProject, SRA, and Bio-Sample numbers are PRJNA680208, SAMN16872356, and SRR13107963, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative genomics and phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:1–12.
- Mehmood F, Abdullah Ubaid Z, Bao YM, Poczai P, Mirza B. 2020. Comparative plastomics of ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9:752.
- Mehmood F, Abdullah Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 8:e9552.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–235.
- Van Tuyl JM, van Creij MGM. 2007. Tulip. In: Anderson NO, editor. Flower breeding and genetics. Dordrecht: Springer; p. 623–641.
- Xing GM, Qu LW, Zhang YQ, Xue L, Su JW, Lei JJ. 2017. Collection and evaluation of wild tulip (Tulipa spp.) resources in China. Genet Resour Crop Evol. 64(4):641–652.
- Xing GM, Qu LW, Zhang W, Zhang YQ, Yuan XF, Lei JJ. 2020. Study on interspecific hybridization between tulip cultivars and wild species native to China. Euphytica. 216(4):66.