Abstract
As a kind of rare plant with important medicinal value and breeding value, Rohdea wattii usually grow in dark and damp places. In this study, we reported the complete chloroplast genome of R. wattii. We used a variety of bioinformatics analysis methods to analyze and visualize the chloroplast genome and phylogenetic relationships. A typical quadripartite structure was observed in the chloroplast genome of R. wattii with a genome size of 156,487 bp. The length of the large single-copy region (LSC) was 84,969 bp, which was the longest region among the four regions; the second longest was the Inverted repeat region (IR) with 26,451 bp in length; and the smallest was the small single-copy region (SSC) with 18,616 bp in length. The overall GC content of R. wattii chloroplast genome is 37.61%. In addition, a total of 132 genes were identified in the whole genome of R. wattii, which include 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Finally, by constructing a phylogenetic tree to analyze the phylogeny of R. wattii, and it indicated that R. wattii and Rohdea chinensis are a close evolutionary relationship.
The Rohdea wattii is a perennial herb, which belongs to the family Asparagaceae (Yamashita and Tamura Citation2004; Nguyen et al. Citation2021). Rohdea wattii grows in dark and humid places and often appears in China, Vietnam, and other places (Tanaka Citation2010). As a kind of medicinal plant, R. wattii has long been used by the folk to treat sore throat, and it also has the medicinal effects of clearing heat and detoxification, dispelling blood stasis, and relieving pain (Shen et al. Citation2003). As a kind of plant with high medicinal value, R. wattii has been widely studied in the fields of pharmacology and phytochemistry (Yao et al. Citation2021), but the analysis of its genetic information is still scarce. We believe it to be an important yet underappreciated direction for the taxonomy and identification of this species. As the wild resources of R. wattii are gradually decreasing (Shen et al. Citation2003), it is more and more urgent to study its genetic and evolutionary relationship at the molecular level. In this study, we used Illumina high-throughput sequencing technique to obtain the complete chloroplast genome sequence of R. wattii, and analyzed its structural characteristics and phylogenetic relationship. These valuable results can facilitate us to efficiently formulate a strategy for the conservation of plant genetic resources, and minimize the risk of extinction of endangered plants.
In this study, the total genomic DNA of R. wattii was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987) and then quantified using a Nanodrop 2000 spectrophotometer. The samples were collected from Guangxi Institute of Botany, Chinese Academy of Sciences, Guilin, China (25°01′N, 110°17′E). Voucher specimen of R. wattii was deposited at the herbarium of Guangxi Institute of Botany (contact person: Bo Zhao, email: [email protected]) under the voucher number ZB-RW202006003).
The sequencing was carried out on the Illumina Novaseq platform following the manufacturer’s protocol (Illumina, San Diego, CA, USA). After quality assessment, the chloroplast genome related reads were filtered by mapping all the raw reads to the reference chloroplast genome of Rohdea chinensis (MH356725; Zhou et al. Citation2018). The chloroplast genome annotation was performed through the online program CPGAVAS (Liu et al. Citation2012) and GeSeq (Tillich et al. Citation2017), and we manually corrected some necessary genes. Then the circular genome map was drawn by the OGDRAW program (Greiner et al. Citation2019) and the whole chloroplast genome of R. wattii was submitted to GenBank (MW822041). And the associated BioProject, SRA, and Bio-Sample numbers are PRJNA720828, SRR14203200, and SAMN18680948, respectively.
Like typical angiosperms (Wicke et al. Citation2011), the chloroplast genome of R. wattii possessed a characteristic quadripartite circular which consists of four regions: LSC, SSC and a pair of IRs. The total length of R. wattii chloroplast genome was 156,487 bp. The large single-copy region (LSC) in R. wattii was 84,969 bp and the small single-copy region (SSC) was 18,616 bp, which were separated by a pair of 26,451 bp inverted repeat regions (IRs). In the complete chloroplast genome, the contents of adenine (A), thymine (T), guanine (G), and cytosine (C) are 30.87%, 31.52%, 18.44%, and 19.17%, respectively. In addition, the GC content of IR region was 42.97%, which was the highest among the four regions; the second highest was LSC region, which was 35.61%; and the GC content in the SSC region was the lowest at 31.49%. A total of 132 genes were identified in the whole genome of R. wattii, including 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Among them, there were 44 genes involved in photosynthesis, which included five genes for photosystem I, 15 genes for photosystem II, six genes for ATP synthase, 11 genes for NADH dehydrogenase, six genes for the cytochrome b/f complex, and one gene for the large chain of Rubisco. These genes related to photosynthesis are the key genes for studying the genetic evolution of R. wattii.
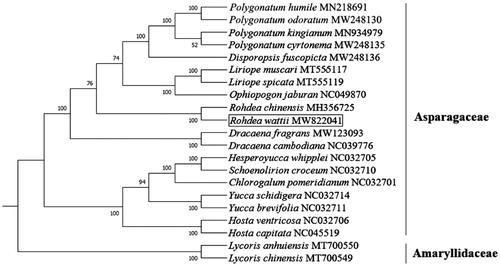
To study the phylogenetic relationship of R. wattii, a phylogenetic analysis was performed based on the complete chloroplast genome sequences of 19 species including the R. wattii chloroplast genome assembled in this study from Asparagaceae and two species from other families. The chloroplast genomes of 20 species were downloaded from the NCBI GenBank database. The sequences were aligned using MAFFT v7 (Mayor et al. Citation2000). Then we constructed the phylogenetic tree using Maximum Likelihood (ML) method by RAxML 7.0.4 (Stamatakis Citation2006) with 1000 replicates under the GTR + CAT model. Lycoris anhuiensis and Lycoris chinensis were designated as outgroup species. The phylogenetic tree revealed that R. wattii was a sister to Rohdea chinensis, and they contributed to a new evolutionary relationship to R. wattii in Rohdea, which is a genus of Asparagaceae in the order Asparagales (). This analysis will help us to understand the evolutionary history of Asparagaceae species and provide new insights for better protection of endangered species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The associated BioProject, SRA, and Bio-Sample numbers are PRJNA720828, SRR14203200, and SAMN18680948, respectively.
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW822041.
Tree file of 21 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.14609466.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715–717.
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 16(11):1046–1047.
- Nguyen KS, Tanaka N, Averyanov LV, Nguyen PH, Tran DB. 2021. Rohdea dangii (Asparagaceae), a new species from northwestern Vietnam. Phytotaxa. 482(1):65–72.
- Shen P, Wang SL, Yang CR, Cai B, Yao XS. 2003. Polyhydroxylated steroidal sapogenins from tupistra wattii. Acta Botanica Sinica. 45(5):626–629.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tanaka N. 2010. A taxonomic revision of the genus Rohdea (Asparagaceae). Makinoa New Series. 9:1–54.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Wicke S, Schneeweiss GM, DePamphilis CW, Kai FM, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.
- Yamashita J, Tamura MN. 2004. Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato), with some taxonomic treatments. J Plant Res. 117(5):363–370.
- Yao SF, Zhang HB, Cai ZS, Gao H, Wang D, Shang SB. 2021. Two new steroidal sapogenins from Rohdea chinensis (synonym Tupistra chinensis) rhizomes and their antifungal activity. Journal of Asian Natural Products Research. 1:1–10.
- Zhou Y, Wang Y, Shi X, Mao S. 2018. The complete chloroplast genome sequence of Campylandra chinensis (Liliaceae). Mitochondrial DNA B Resour. 3(2):780–781.