Abstract
Ilex rotunda is a traditional Chinese medicine plant. In this study, we characterized the complete chloroplast (cp) genome sequence of I. rotunda to investigate its phylogenetic relationship. The cp genome of I. rotunda was 157,743 bp in length with 37.62% overall GC content, including a large single-copy (LSC) region of 87,060bp, a small single-copy (SSC) region of 18,432 bp, which were separated by a pair of inverted repeats (IRS) of 26,121 bp. The cp genome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on whole cp genome sequences showed that I. rotunda is closely related to I. pubescens and I. polyneura.
Introduction
Ilex rotunda Thunberg 1784 belongs to Ilex Linnaeus which is within the monogeneric family, i.e. Aquifoliaceae, that includes ca. 700 species (Yao et al. Citation2021). Ilex have two diversity centers, South East Asia-Malesia and South America, but few species in Europe and Africa. China is one of the Ilex diversity centers that there are 200 species recognized (Xie et al. Citation2019). In China, more than 40 species and their varieties, including I. rotunda, are recorded for the medicinal purpose (Hao et al. Citation2015). In terms of medicinal value, Ilex is effective to clearing away heat and detoxification, detumescence and pain relief. Accordingly, China is one of the major countries that have abundant natural Ilex resources. However, the morphological study based on leaves, inflorescence, and fruit is difficult to distinguish close relative species within Ilex. The molecular phylogeny methods can be a useful tool for systematics study. The complete chloroplast genome can provide a framework to insight Ilex phylogenetic relationship (Nei and Kumar Citation2000). Ilex rotunda is widely distributed is southern China, that can be used for traditional Chinese medicine and for garden plant.
I. rotunda was planted in Nanjing Botanical Garden, Mem. Sun Yat-sen (118°49′55″E, 32°3′32″N), Nanjing, China. The voucher specimen (NO.NBGJIB-Ilex-0056) was deposited in the Institute of Botany, Jiangsu Province and Chinese Academy of Science (http://www.cnbg.net/, hongchen, [email protected]). Approximately 3 g of fresh leaves were collected and stored in liquid nitrogen for chloroplast DNA isolation. The qualified DNA was used for library construction and sequencing (Illumina, San Diego, CA). In total, 6710.3 Mb of raw data and 6352.4 Mb clean data were obtained. De novo genome assembly and annotation were conducted by the organelle assembler NOVOPlasty (Dierckxsens et al. Citation2017) and GeSeq (Tillich et al. Citation2017), respectively. The complete cp genome was deposited in GenBank (accession number: MW292559).
The shape of the cp genome of I. rotunda was a double-stranded closed loop. The cp genome of I. rotunda was 157,743 bp in length, including a large single-copy (LSC) region of 87,060 bp, a small single-copy (SSC) region of 18,432 bp. The cp genome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. For the 132 identified genes, 14 contained introns, 4 (rps12, ycf3, rps12, clpP) had two introns and the rests had a single intron. The overall GC content of the circular genome was 37.62%.
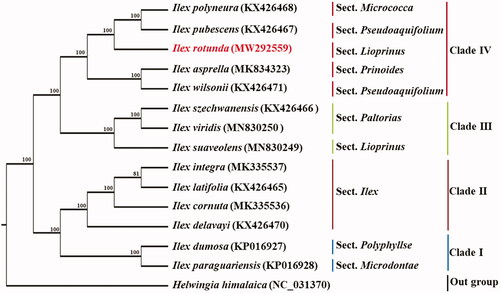
To explore the evolution status of Ilex rotunda, 14 other complete chloroplast genome sequences were downloaded from the Genebank database (Helwingia himalaica as outgroup). Sequences alignment was firstly performed by MAFFT (Katoh and Standley Citation2013), and then phylogenetic tree was constructed with maximum-likelihood method, using the IQ-tree program (Nguyen et al. Citation2015). Our phylogenetic analysis indicated that all the examined Ilex species were divided into four clades, and I. rotunda has a close relationship with I. pubescens and I. polyneura (). The work herein provided a better understanding of I. rotunda, and plays a critical role in constructing phylogeny of Aquifoliaceae.
Figure 1. Phylogenetic tree based on the cp genome sequences of Ilex rotunda and 13 other species (contain 1 outgroup). Section names were displayed in the right side of phylogenetic tree (Su et al. Citation2020). Numbers on the nodes indicate bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that obtained at this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession number of MW292559. The associated BioProject, Bio-Sample and SRA, numbers are PRJNA747869, SAMN20298920 and SRR15195658, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hao DC, Gu XJ, Xiao PG. 2015. Phytochemistry and biology of Ilex pharmaceutical resources. Med Plants. 2015:531–585.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Su T, Zhang M, Shan Z, Li X, Zhou B, Wu H, Han M. 2020. Comparative survey of morphological variations and plastid genome sequencing reveals phylogenetic divergence between four endemic Ilex species. Forests. 11(9):964.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Xie Y, Wu F, Fang X. 2019. Middle Eocene East Asian monsoon prevalence over southern China: evidence from palynological records. Glob Planet Change. 175:13–26.
- Yao X, Song Y, Yang JB, Tan YH, Corlett RT. 2021. Phylogeny and biogeography of the hollies (Ilex L., Aquifoliaceae). J Syst Evol. 59(1):73–82.
