Abstract
The mitochondrial genome of Hemathlophorus brevigenatus Wei, 2005 collected from Huanggang Mountain of China is described using the NGS approach. The length of the sequence is 15,452 bp containing 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one control region. The overall A + T content is 79.5%. tRNA rearrangements occur in the MQI cluster. Phylogenetic analysis of H. brevigenatus resolved it in a clade with Allantus togatus in Allantinae which provides new evidence for the phylogeny of Tenthredinidae.
Lacourt (Citation1996) erected the tribe Athlophorini for Hemathlophorus Malaise, Citation1945 and Athlophorus Burmeister, 1847 under the subfamily Sioblinae of Tenthredinidae. Although Hemathlophorus and Athlophorus are two peculiar genera among the Tenthredinidae as shown by the fore wing with a long vein R + M and a more or less basally constricted abdomen, Lacourt’s peculiar systematic arrangement of the genera and the tribe was not adopted by sawfly researchers, such as Wei and Nie (Citation1998), Saini (Citation2006), and Taeger et al. (Citation2010). In the three later systems, Hemathlophorus and Athlophorus were placed into the Allantinae of Tenthredinidae. In this study, we sequenced the mitochondrial genome of Hemathlophorus brevigenatus Wei, Citation2005 and inferred its phylogenetic history with other mitochondrial genomes of Symphytan species to clarify the phylogenetic position of Hemathlophorus within the Tenthredinidae.
Specimens of H. brevigenatus were collected from Huanggangshan Mountain, Fujian Province, China (27.81 N, 117.95 E) in April 2019. The specimen was deposited at the Asia Sawfly Museum, Nanchang (ASMN) (Meicai Wei, [email protected]) under the voucher number CSCS-Hym-MC0177. Whole genomic DNA was extracted from the thorax muscle of a female adult using the DNeasy Blood &Tissue Kit (Qiagen, Valencia, CA). Genomic DNA was analyzed with the high-throughput Illumina Hiseq 4000 platform with 150 bp paired-end reads. DNA sequences were assembled using MitoZ (Meng et al. Citation2019) and Geneious Prime 2019.2.1 (Biomatters Ltd., Auckland, New Zealand). A total of 43,205,384 raw reads were assembled using MitoZ and resulted in a contig 15,108 bp in length with 37 genes. The control region was assembled using trnM, trnI from the Xenapatidea procincta (MW487928) as a reference. The contig was extended 3′ by 117 bp and 5′ by 582 bp using the “Map to Reference” function in Geneious Prime. This extension that resulted overlapped with the 15,108 bp contig and closed the gap. Thus, a 482 bp long control region was obtained. The verification is conducted by assembly using X. procincta as the reference. The annotations of tRNAs were generated using the MITOS web server (Bernt et al. Citation2013). Protein-coding genes (PCGs) were annotated by the open reading frames between the flanking tRNAs, and then defined by comparative analyses (Cameron 2014).
The complete mitochondrial genome sequences were aligned with ClustalW using default settings (Thompson et al. Citation1994) and concatenated with SequenceMatrix v1.7.8 (Vaidya et al. Citation2011). The phylogenetic tree was constructed using IQ-TREE (maximum-likelihood) (Nguyen et al. Citation2015) with the GTR + MTART model and 1000 bootstrap replicates. To investigate the phylogenetic position of H. brevigenatus, 10 unsaturated nucleotide sequences (atp8, nad2, nad6 are excluded) of 37 Hymenopterans are aligned separately then concatenated, resulting in an alignment of 9836 bp.
The total length of the complete mitochondrial genome is 15,452 bp, containing 13 PCGs, 22 transfer RNA genes, two ribosomal RNA genes, and one control region. The mitochondrial base composition is A 42.5%, T 37.0%, G 7.8%, and C 12.7%. The AT content is 79.5% and is common in the Tenthredinidae (Ma et al. Citation2019). All of the PCGs are initiated with the ATN (ATT, ATA, and ATG) codon, except nad4l which initiated from GTG. Among these genes, three PCGs (nad1, nad5, and nad6) initiated with ATA, seven PCGs (atp6, cob, cox1, cox2, cox3, nad2, and nad4) with ATG and two PCGs (atp8 and nad3) initiated from ATT. All the PCGs have a TAA termination codon except nad4 which has an incomplete terminal codon T—. Compared with the ancestral insect (Drosophila) mitochondrial genome (Boore Citation1999), the control region- trnI (+)- trnQ (-)- trnM (+) is rearranged to trnM (-)- trnQ (+)- control region- trnI (+) in H. brevigenatus. There are five gene overlapping regions that appeared among trnI-nad2 (1 bp), trnW-trnC (1 bp), atp6-atp8 (7 bp), atp6-cox3 (1 bp), and nad6-cob (1 bp). The mitogenome has nine intergenic spacers with a total length of 173 bp in 18 locations varying in size from 1 to 39 bp with the longest located between trnF and nad5.
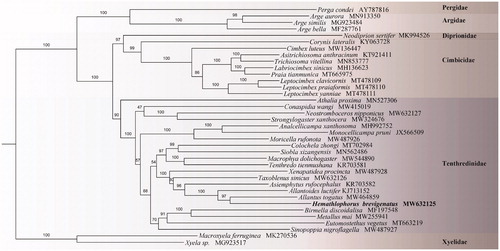
Phylogenetic analysis of H. brevigenatus fully resolved it in a clade with Allantus togatus (MW464859) and Allantoides luctifer (KJ713152) classified in the Allantinae as suggested by Wei and Nie (Citation1998) and Taeger et al. (Citation2010). Its placement was remote from the Siobla of Tenthredininae and clearly denied Lacourt’s system (Lacourt Citation1996). The phylogenetic relationships of Tenthredininae are inferred as (Athalia + ((Conaspidia + Selandriinae) ((Hoplocampinae + Nematinae) ((Tenthredininae + Allantinae) + ((Fenusinae + Blennocampinae) + Caliroinae))))) (). All related files are publicly available in figshare (https://figshare.com/account/home#/projects/114354).
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number MW632125. The associated BioProject, SRA, and BioSample numbers are PRJNA714776, SRR14233978, and SAMN18397095 respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cameron S. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):400–411.
- Lacourt J. 1996. Contribution à une révision mondiale de la sous-famille des Tenthredininae (Hymenoptera: Tenthredinidae). Annales de la Société Entomologique de France. N. S. 32(4):363–402.
- Ma Y, Zheng BY, Zhu JC, Achterberg C, Tang P, Chen XX. 2019. The first two mitochondrial genomes of wood wasps (Hymenoptera: Symphyta): Novel gene rearrangements and higher-level phylogeny of the basal hymenopterans. Int J Biol Macromol. 123:1189–1196.
- Malaise R. 1945. Tenthredinoidea of South-Eastern Asia with a general zoogeographical review. Opuscula Entomologica Suppl. 4:1–288.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von HA, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Saini MS. 2006. Subfamilies Allantinae. In: Indian sawflies biodiversity. Keys, catalogue & illustrations. Vol. III. Dehra Dun: Bishen Singh Mahendra Pal Singh; p. 1–182.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Taeger A, Blank SM, Liston AD. 2010. World catalog of Symphyta (Hymenoptera). Zootaxa. 2580(1):1–1064.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Wei M. 2005. On the sawfly genus Hemathlophorus Malaise of China (Hymenoptera, Tenthredinidae, Allantinae). Acta Zootaxonomica Sinica. 30(4):822–827.
- Wei M, Nie H. 1998. Generic list of Tenthredinoidea s. str. (Hymenoptera) in new systematic arrangement with synonyms and distribution data. J Central South For Univ. 18(3):23–31.