Abstract
Cinnamomum septentrionale is a commercially important timber tree and wild spice plant classified in the Lauraceae. Here we sequenced the complete plastid genome of C. septentrionale to contribute to its phylogenetics and bioinformatics. The chloroplast genome of C. septentrionale is 152,729 bp in length and includes a large single copy (LSC) region of 93,639 bp, a small single copy (SSC) region of 18,858 bp, and two inverted repeat (IR) regions of 20,114 bp. The GC content of LSC, SSC, IR and the whole genome is 37.9%, 33.8%, 44.4%, and 39.1%, respectively. There are 128 genes, including 84 protein-coding, 36 tRNA, and eight rRNA genes. The phylogenetic analysis resolved C. septentrionale in a sister position to C. glanduliferum. The complete chloroplast genome sequence of C. septentrionale provides valuable genomic information for future systematic studies on Cinnamomum.
Cinnamomum septentrionale Hand-Mazz, a large-sized evergreen tree species, is widely distributed in Guizhou, Sichuan, and Yunnan of SW China (http://foc.iplant.cn/). The essential oil isolated from the bark and leaves of C. septentrionale is rich in eucalyptol (Taha and Eldahshan Citation2017) and thus represent an important aromatic plant species in the genus Cinnamomum Trew (Kumar and Kumari Citation2019). For a better understanding of the relationships of C. septentrionale and other Cinnamomum species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches. In the present study, we report and characterize the complete chloroplast (cp) genome of C. septentrionale based on Illumina pair-end sequencing and compare it to other genome sequences from Lauraceae. The results provide valuable genetic information for evolutionary and conservation studies of C. septentrionale.
The voucher specimen of C. septentrionale was collected from Nanchong, Sichuan province, China (106°08′E; 30°79′N) and is deposited at the herbarium of NanYang Medical College, Department of Chinese Medicine (http://www.nymc.edu.cn/, Lv Qin, [email protected]) under the voucher number YM001. The DNA sample is stored at the Key Laboratory of Department of Chinese Medicine, Nanyang, China. Total genomic DNA was extracted using the DNA Secure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol. Library preparation and genomic sequencing on the Illumina Hiseq 2500 platform were conducted by Benagen (Benagen Inc., Wuhan, China). The raw sequence data have been deposited into NCBI SRA with project accession of SRR14793489. The raw data was filtered using Trimmomatic Version 0.32 with the default settings (Bolger et al. Citation2014). The filtered output yielded 5.4 Gb of raw 150 bp paired-end data. The obtained paired-end reads were assembled using SPAdes v.3.9.0 (Nurk et al. Citation2017). The assembled sequence was annotated in MPI-MP CHLOROBOX (https://chlorobox.mpimp-golm.mpg.de/geseq.html) via GeSeq using the reference cp genome of C. glanduliferum (MW369062, Zhao et al. Citation2019), and corrected using Geneious Prime v2020.2 (Kearse et al. Citation2012). Finally, the complete chloroplast genome of C. septentrionale was submitted to GenBank (Accession No. MZ128522).
The chloroplast genome of C. septentrionale is 152,729 bp in length and includes a large single copy (LSC) region of 93,639 bp, a small single copy (SSC) region of 18,858 bp, and two inverted repeat (IR) regions of 20,114 bp. The GC contents of LSC, SSC, IR and whole genome are 37.9%, 33.8%, 44.4%, and 39.1%, respectively. The chloroplast genome encodes 128 genes, including 84 protein-coding, 36 tRNA, and eight rRNA genes. The plastome of C. septentrionale was 14 bp and 10 bp larger than that of C. glanduliferum (MW369062) and C. bodinieri (152,719 bp, MH394415). It was also 31 bp and 46 bp smaller than that of C. parthenoxylon (152,760 bp, MH050971) and C. burmanni (152,775 bp, LAU00110). The overall GC content of C. septentrionale is consistent with C. glanduliferum (MW369062), C. bodinieri (MH394415) and C. parthenoxylon (MH050971), which are all 39.1%.
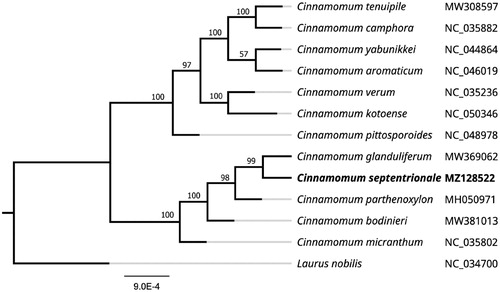
To further investigate the systematics of C. septentrionale, a Maximum-likelihood (ML) tree was constructed using complete chloroplast genome sequences of 12 other related species aligned with MAFFT (Katoh and Standley Citation2013). The analysis was performed using MEGA 7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The program operating parameters were set as follows: a Tamura 3-parameter (T92) nucleotide substitution model with 1000 bootstrap repetitions, accompanied by Gamma distributed with Invariant site (G + I) rates, and partial deletion of gaps/missing data. The phylogenetic analysis revealed that C. septentrionale was most related to C. glanduliferum as a sister group with 99% bootstrap support. Lauraceae are an important component of tropical and subtropical forests and have major ecological and economic significance. Owing to lack of clear-cut morphological differences between genera and species, this family is an ideal case for testing the efficacy of DNA barcoding in the identification and discrimination of species and genera. The NJ tree based on the combined barcodes rbcL + matK + trnH–psbA + ITS revealed that C. glanduliferum was most related to C. parthenoxylon as a sister group owing to the previous studies did not include C. septentrionale (Liu et al. Citation2017). The complete chloroplast genome sequences of C. septentrionale will provide valuable bioinformatic information for future phylogenetic studies and the systematics of Cinnamomum ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. MZ128522. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA737038, SRR14793489, and SAMN19678362 respectively.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Zhao G, Yang J, Wang X, Song Y, Zhu R. 2019. The plastid genome of a spice plants Cinnamomum glanduliferum in Tibet (Lauraceae). Mitochondrial DNA B Resour. 4(2):3284–3285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Kumari R. 2019. Cinnamomum: review article of essential oil compounds, ethnobotany, antifungal and antibacterial effects. Open Access J Sci. 3:13–16.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu Z-F, Ci X-Q, Li L, Li H-W, Conran JG, Li J. 2017. DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PLoS One. 12(4):e0175788.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Taha MA, Eldahshan AO. 2017. Chemical characteristics, antimicrobial, and cytotoxic activities of the essential oil of Egyptian Cinnamomum glanduliferum bark. Chem Biodiversity. 14(5):e1600443.