Abstract
Alnus formosana (Betulaceae) is an important ecological and economic deciduous tree species widely distributed throughout subtropical regions of Taiwan province, China. At the present study, the complete chloroplast genome of A. formosana was assumbled using next-generation sequencing technology. The complete chloroplast sequence is 161,029 bp in length, which consisted of a large single copy (LSC, 89,720 bp) and a small single copy (SSC; 19,205 bp) separated a pair of inverted repeats (IRs; 26,052 bp). The overall guanine-cytosine (GC) content was 36.4%. A total of 131 genes were annotated, including 85 protein-coding genes, 37 tRNAs, eight rRNAs and one pseudogene (ψycf1). The phylogenetic analysis fully resolved A. formosana in a clade with A. japonica. The plastome of A. formosana will provide informative genomic resources for further phylogenetic application and genetic improvement.
Alnus formosana (Burkill) Makino 1912, classified in the birch family (Betulaceae), is an important ecological and economic deciduous tree species(Li and Skvortsov Citation1999). It has a wide natural distribution throughout the subtropical regions in the Taiwan province, China (Liao and Weng Citation2002). It is characterized by fast-growing, nitrogen-fixing and wide ranging adaptability to adverse environmental conditions, and is, therefore, widely used as pioneer tree of afforestation (Pan et al. Citation2008). This species is also a source of excellent quality wood for furniture and industrial production. However, the phylogenetic position of A. formosana within Alnus has not yet been sufficiently resolved (Ren et al. Citation2010). Here, we report the complete chloroplast (cp) genome sequence of A. formosana using next-generation sequencing, which will lay the foundation for further phylogenetic application and genetic resolution.
Total genomic DNA was extracted from the fresh leaves of A. formosana cultivated in Guangxi Forestry Research Institute, Guangxi Province, China (22°55′30″N, 108°21′E) by modified hexadecyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The specimen was deposited at Guangxi Forestry Research Institute (contact: Zihai Qin, [email protected]) under voucher number: 20210303002. The isolated genomic DNA was subsequently sequenced on an Illumina Hiseq X-ten platform (San Diego, USA) at Novogene (Beijing, China). The raw Paired-end (PE) reads were de novo assembled into the chloroplast genome using the perl script NOVOPlasty 4.3.1 (Dierckxsens et al. Citation2017) with default settings. The assembled genome was annotated via GeSeq (Tillich et al. Citation2017) and adjusted manually in Geneious 11.1.5 (Kearse et al. Citation2012).
The chloroplast genome of A. formosana has a circular structure with a length of 161,029 bp, comprising four parts: a LSC (89,720 bp), a SSC (19,205 bp) and a pair of IRs (26,052 bp). The overall guanine-cytosine (GC) content was 36.4%, with the GC contents of LSC, SSC and IRs at 34.1%, 30.1%, and 42.6%, respectively. The complete sequence data was submitted to National Center for Biotechnology Information (NCBI) (accession number MW865380). A total of 131 genes were annotated, including 85 protein-coding, 37 transfer RNA (tRNA), eight ribosomal RNA (rRNA) and one pseudogene (ψycf1). Among them, four rRNA (4.5S, 5S, 16S and 23S rRNA), seven tRNA (trnI-GAU, trnA-UGC, trnL-CAA, trnI-CAT, trnR-ACG, trnV-GAC, trnN-GTT), and seven protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12, and ycf15) were duplicated in the IR regions. The plastome of A. formosana had similar in the terms of structure, gene content and order, and GC content compared to those of other published Alnus.
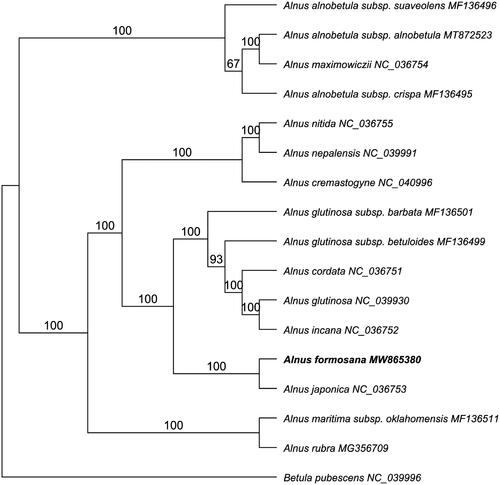
To reveal the phylogenetic relationship of A. formosana within Alnus, we downloaded additional plastomes from NCBI including fifteen Alnus and one Betula species. The whole plastome sequences were aligned by MAFFT 7.409 (Katoh and Standley Citation2013) using default settings. The phylogenetic analysis was generated by the GTR + GAMMA model in RAxML with 1,000 bootstrap replicates (Stamatakis Citation2014). The maximum likelihood (ML) phylogenetic tree indicated that Alnus is a monophyletic group which is consistent with the previous analysis by Chen and Li (Citation2004) but with higher bootstrap support (100%) (). Within genus Alnus, A. formosana is most closely related to A. japonica (NC_036753) with a 100% bootstrap value, which is in accordance with their morphological features (Ren et al. Citation2010).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW865380. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA743224, SRR15021752 and SAMN20003570, respectively.
Additional information
Funding
References
- Chen ZD, Li JH. 2004. Phylogenetics and biogeography of Alnus (Betulaceae) inferred from sequences of nuclear ribosomal DNA ITS region. Int J Plant Sci. 165(2):325–335.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li P, Skvortsov AK. 1999. Betulaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 4. Beijing: Science Press; St. Louis: Missouri Botanical Garden.
- Liao T, Weng J. 2002. Ecophysiological characteristics of Taiwan alder (Alnus formosana) seedlings adapted to the subtropical region. Tree Physiol. 22(5):355–362.
- Pan Y, Li X, Rong L, Yuan W, Wang J. 2008. Root nodulation of young Alnus formosana on forest lands converted from agricultural lands. Chinese J Ecol. 27:1482–1486.
- Ren B, Xiang X, Chen Z. 2010. Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Mol Ecol Resour. 10(4):594–605.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.