Abstract
In this study, the complete chloroplast genome of Loropetalum chinense var. rubrum Yieh was sequenced and characterized. The chloroplast genome is 159,427 bp in length, and contains a large single-copy (LSC) region of 88,185 bp and a small single-copy (SSC) region of 18,772 bp separated by a pair of inverted repeat (IR) regions of 26,235 bp. There are 128 genes in the genome including 83 protein-coding, 37 tRNA, and eight rRNA genes. The overall GC is 38.0% and the LSC, SSC, and IR regions are 36.1, 32.7, and 43.1%, respectively. Phylogenetic analysis of L. chinense var. rubrum fully resolved it in a clade with L. subcordatum. This complete chloroplast genome will provide valuable insight into evolution, molecular breeding, and phylogenetic analysis of Loropetalum species.
Loropetalum chinense var. rubrum is a species classified in the Hamamelidaceae, a shrub mainly distributed in China, and India (Zhang et al. Citation2020). It shares a vast application in city landscaping due to its colorful flowers and bright leaves (Li et al. Citation2020). This species is also used as an important traditional medicinal in China, and has a high medicinal value (Tang et al. Citation2011).
Chloroplast genome in plants is exceptionally conserved in gene content and organization (Moore et al. Citation2010); therefore, they are ideal for ecological, evolutionary, and diversity studies (Wicke et al. Citation2011). At present, the complete chloroplast genomes of several species from the Hamamelidaceae have been published (Kim et al. Citation2019; Zhang et al. Citation2019). However, the chloroplast genome of L. chinense var. rubrum has not been investigated. Here, we determined the complete chloroplast genome sequence of L. chinense var. rubrum to contribute to the bioinformatics and systematics of the Hamamelidaceae and to document the genetic structure and content of this species.
The fresh leaves were collected from Tropical and Subtropical Economic Crops Institute of Yunnan Academy of Agricultural Science, Baoshan, China (Yunnan, China; geospatial coordinates: 99°10′25.61″E, 25°08′0.11″N; altitude: 1655 m). The voucher specimen of L. chinense var. rubrum was deposited at the herbarium of Yunnan Academy of Agricultural Sciences, Kunming, China under the voucher number YAAS-JM-006 (url: http://www.yaas.org.cn/, contact person: Mr Mingkun Xiao, and email: [email protected]) and DNA samples were stored at the Laboratory of Yunnan Academy of Agricultural Sciences, Kunming, China. The total genomic DNA was extracted using the Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China). We assembled the chloroplast genome using GetOrganelle v1.6.2e (Jin et al. Citation2020). The annotation was performed with the online annotation tool CPGAVAS2 (Shi et al. Citation2019). The annotated chloroplast is deposited in GenBank under accession number MW368385.
The complete chloroplast genome of L. chinense var. rubrum is 159,427 bp in length, and contains a pair of inverted repeat (IR) regions of 26,235 bp, a large single-copy (LSC) region of 88,185 bp and a small single-copy (SSC) region of 18,772 bp. The overall GC content is 38.0%, the corresponding values of the LSC, SSC, and IR region are 36.1, 32.7, and 43.1%, respectively. There are 128 genes, including 83 protein-coding, eight rRNA, and 37 tRNA genes. Among them 16 genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnA-UGC, trnI-GAU, rps16, rpoC1, rpl16, rpl2, rpl23, petB, petD, ycf2, ndhB, ndhA) have single introns, while two genes (ycf3 and clpP) have double introns. The chloroplast of this species was very similar to that of other species of the Hamamelidaceae, such as Loropetalum subcordatum (Zhang et al. Citation2019) and Corylopsis spicata (Kim et al. Citation2019).
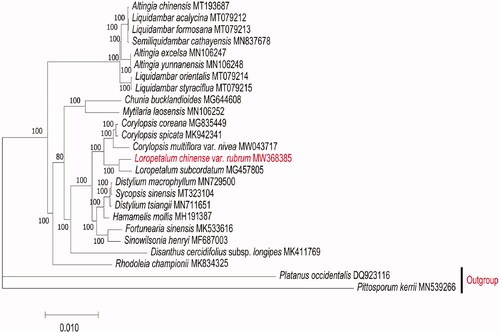
Phylogenetic analysis was based on the complete chloroplast genome of L. chinense var. rubrum, together with other 24 species of Hamamelidaceae downloaded from GenBank. The alignment was performed using the default settings in MAFFT v7.471 (Katoh et al. Citation2019) and analyzed by IQ-TREE v1.6.12 (Minh et al. Citation2020) under the TVM + F+R3 nucleotide substitution model and 1000 bootstrap replicates. The phylogenetic tree suggested that, L. chinense var. rubrum and L. subcordatum were clustered together, and shared a closer relationship to Corylopsis species (), which is consistent with the previous phylogenetic studies (Kim et al. Citation2019; Chen et al. Citation2020; Lv et al. Citation2021). The complete chloroplast genome of L. chinense var. rubrum provides essential data for further biological, bioinformatic, and systematic analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number: MW368385. The associated Bio-Project, Bio-Sample, and SRA numbers are PRJNA689662, SAMN17213685, and SRR13356418, respectively.
Additional information
Funding
References
- Chen SF, Li Y, Wang L, Xie L, Ge XM, Zhou X, Hu YP, Zhang WW, Fang YM, Ding H. 2020. The complete chloroplast genome sequence of Distylium macrophyllum (Hamamelidaceae). Mitochondrial DNA B Resour. 5(1):701–702.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kim SC, Shin SY, Ahn JY, Lee JW. 2019. Complete chloroplast genome of Corylopsis spicata and phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2700–2701.
- Li CH, Rong DY, Liao XS, Zhang BY. 2020. Cloning and bioinformatic analysis of two chalcone synthases from Loropetalum chinense var. rubrum. J Hunan Univ Technol. 34(6):92–98.
- Lv T, Chen SF, Zhao R, Wang NJ, Fang YM. 2021. The complete chloroplast genome sequence of Corylopsis multiflora Hance var. nivea Chang. Mitochondrial DNA B Resour. 6(1):271–273.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von HA, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci U S A. 107(10):4623–4628.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tang H, Zheng QF, Ge G, Xue XW, Yan AP, Peng L, Sun BT. 2011. Analysis of fatty acids in leaves of Loropetalum chinense and L. chinense var. rubrum by GC–MS. Zhong Yao Cai. 34(10):1549–1552.
- Wicke S, Schneeweiss GM, Depamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.
- Zhang BY, Li CH, Liu X, Liao XS, Rong DY. 2020. Cloning and subcellular localization analysis of LcDFR1 and LcDFR2 in Loropetalum chinense var. rubrum. J South Agric. 51(12):2865–2874.
- Zhang YY, Cai HX, Dong JX, Gong W, Li P, Wang ZS. 2019. The complete chloroplast genome of Loropetalum subcordatum, a national key protected species in China. Conserv Genet Resour. 11(4):377–380.