Abstract
Gymnocypris dobula, classified into the highly specialized Schizothoracine fish, is endemic to Tibet, China. The complete mitochondrial DNA sequence of G. dobula was 16,728 base pairs in length and comprised 22 transfer RNA genes, 13 protein-coding genes, two ribosomal RNA genes as well as one control region as in a typical vertebrate mitochondrial DNA gene. The ML and BI trees showed that G. dobula was most closely related to Gymnocypris scleracanthus within the highly specialized group. This mitogenome provides new molecular data for further taxonomic and phylogenetic studies of Schizothoracine fish.
The Schizothoracine fish, one of three broad fish lineages (including the Glyptosternoids and Triplophysa) generally discovered on the Qinghai-Tibetan Plateau, plays an essential ecological role in the plateau ecosystem (Ma et al. Citation2015). Gymnocypris dobula (Cypriniformes: Cyprinidae) is endemic to Tibet, China, and has a high economic value to marginal fishermen (Chan et al. Citation2016). The unique climatic and geographical characteristics of the Qinghai-Tibet Plateau resulted in the complex phylogenetic relationship among the schizothoracinae fish (Liang et al. Citation2017; Quan et al. Citation2021). Based on its morphological characters without any scales and barbels, G. dobula was classified into the highly specialized group of Schizothoracine fish (Qi et al. Citation2012). Owing to its distribution only in several localities, G. dobula is assessed as Vulnerable (VU) status by International Union for Conservation of Nature (IUCN). In this study, the complete mitochondrial DNA sequence of G. dobula was reported contributing to a better understanding of its further genetic studies, and also providing significant information for the reference of systematics and conservation.
The G. dobula sample was collected from Yanghu Lake (N 29.00°, E 90.41°), Tibet, Chian, and was stored in National Engineering Research Center for Marine Aquaculture, Zhejiang Ocean University (Jian, Chen and [email protected]) under the voucher number F20200113. Genomic DNA was extracted from muscle using the standard phenol/chloroform extraction method (Sambrook and Russell Citation2006). The mitochondrial sequences, amplified by PCR with seventeen pairs of primer (Table S1), were obtained through Sanger dideoxy sequencing and assembled by CodonCode Aligner 5.1.5. The assembled mitochondrial genome was annotated using the online tool MITOS (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and software Sequin (version 13.70, http://www.ncbi.nlm.nih.gov/Sequin). The annotated sequence was deposited in GenBank with the accession number MW924117.
Similar to the typical mitogenome of vertebrates, the mitogenome of G. dobula was a closed double-stranded circular molecule of 16,728 bp, including 13 protein-coding genes (PCGs), 22 tRNA genes, two ribosomal RNA genes (12S and 16S rRNA), and a control region (Boore Citation1999; Zhang et al. Citation2017). Most mitochondrial genes, including 12 PCGs, 14 tRNAs, and two rRNAs were encoded on the H-strand except for ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, SerUCA, Glu, and Pro), which are encoded on the L-strand. The overall base composition was A (28.46%), T (27.21%), C (26.06%), G (18.27%), respectively, which showed a negative GC-skew value (–0.176) and positive AT-skew value (+0.023). The nucleotide composition of the whole mitogenome was A + T biased (55.67%), and the A + T content of PCGs, tRNAs, and rRNAs was 56.12%, 54.55%, and 53.60%, respectively. As with other vertebrate mitogenomes, most of these PCGs started by ATG initiation codon, except for COI by GTG (Zhang et al. Citation2020a). As for the stop codon, seven PCGs performed the routine termination codon (TAA or TAG), whereas five other PCGs (ND2, COII, ND3, ND4, and Cyt b) stoped with an incomplete stop codon T and one PCG (COIII) stoped with an incomplete stop codon TA. All of the tRNAs were predicted to be folded into canonical cloverleaf secondary structures except for tRNA-SerAGC using the online tool tRNAscan-SE (Lowe and Chan Citation2016). The 12S rRNA and 16 rRNA genes were 960 bp and 1,682 bp, respectively, which were typically separated by tRNA-Val. The length of the control region was 938 bp, with highly A + T (63.11%) rich.
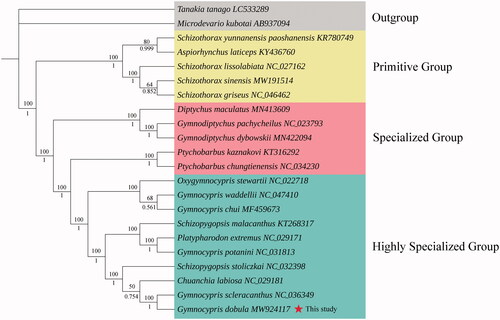
Based on the sequences of 13 PCGs including 20 species of Schizothoracine fish and two outgroup species, we constructed maximum likelihood (ML) and Bayesian inference (BI) phylogenetic trees. The ML and BI trees were constructed by the software PhyML 3.0 (Guindon et al. Citation2010) and MrBayes 3.2.6 (Ronquist et al. Citation2012), respectively, with GTR + F + I + G4 as the best-fit evolutionary model determined by ModelFinder (Kalyaanamoorthy et al. Citation2017). The two trees showed the identical topology structure and indicated that G. dobula was closely related to Gymnocypris scleracanthus (). The genus Gymnocypris was polyphyletic and clustered into the highly specialized group, which was consistent with previous studies (An et al. Citation2020; Zhang et al. Citation2020b).
Figure 1. Phylogenetic analysis based on the sequences of the 13 PCGs in the mitogenome. ML tree with bootstrap values (above, with 100,000 replications) and BI posterior probabilities (below, with 100,000 generations) were shown next to nodes. The number after the species name was the GenBank accession number. The genome sequence in this study is labeled with a red star.
Supplemental Material
Download MS Word (20.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/ MW924117) under the accession number: MW924117.
Additional information
Funding
References
- An Z, Feng S, Wang Y, Li Y, Deng J, Wang W. 2020. Complete mitochondrial genome of Gymnocypris waddellii (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 5(2):1874–1875.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chan J, Li W, Hu X, Liu Y, Xu Q. 2016. Genetic diversity and population structure analysis of Qinghai-Tibetan plateau schizothoracine fish (Gymnocypris dobula) based on mtDNA D-loop sequences. Biochem Syst Ecol. 69:152–160.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Liang Y, He D, Jia Y, Sun H, Chen Y. 2017. Phylogeographic studies of schizothoracine fishes on the central Qinghai-Tibet Plateau reveal the highest known glacial microrefugia. Sci Rep. 7(1):10911–10983.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Ma X, Kang J, Chen W, Zhou C, He S. 2015. Biogeographic history and high-elevation adaptations inferred from the mitochondrial genome of Glyptosternoid fishes (Sisoridae, Siluriformes) from the southeastern Tibetan Plateau. BMC Evol Biol. 15:212–233.
- Qi D, Chao Y, Guo S, Zhao L, Li T, Wei F, Zhao X. 2012. Convergent, parallel and correlated evolution of trophic morphologies in the subfamily schizothoracinae from the Qinghai-Tibetan plateau. PLoS One. 7(3):e34070.
- Quan J, Zhao G, Li L, Zhang J, Luo Z, Kang Y, Liu Z. 2021. Phylogeny and genetic diversity reveal the influence of Qinghai-Tibet Plateau uplift on the divergence and distribution of Gymnocypris species. Aquat Sci. 83(1):1–11.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 2006:pdb.prot4455.
- Zhang C, Liu J, Wang W, Zhou J, Pan Y, Mou Z. 2017. The complete mitochondrial genome of the Gymnocypris scleracanthus (Cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 2(2):634–635.
- Zhang K, Liu Y, Yin X, Yuan P, Chen J, Gao Y, Ping H, Zhang H, Miao Z, Liu B, et al. 2020a. Characterization of the complete mitochondrial genome of Chinese Konosirus punctatus (Clupeiformes, Clupeidae) and phylogenetic studies of Clupeiformes. Mitochondrial DNA B Resour. 5(3):3371–3373.
- Zhang Y, Li X-h, Tian F, Liu S-j, Feng C-g, Zhao K. 2020b. Mitochondrial genome and phylogenetic relationship of Gymnocypris eckloni (Schizothoracinae) in Qaidam river basin. Genomics. 112(6):4316–4321.
