Abstract
The complete sequence of the circular mitochondrial (mt) DNA of Populus rotundifolia var. duclouxiana is presented in this study. The size of the P. rotundifolia var. duclouxiana mt genome was 775,549 bp and the GC content of the genome was 44.8%. The P. rotundifolia var. duclouxiana mt genome encoded 58 genes, including 33 protein-coding genes, 22 tRNA genes, and three rRNA genes. Among the 58 genes, three genes contained one intron, one gene contained three introns, and four genes contained four introns. Phylogenetic analysis using seven complete mt genomes of Salicaceae supported that P. rotundifolia var. duclouxiana is most closely related to P. davidiana.
Populus rotundifolia var. duclouxiana, a variety of P. rotundifolia, is a native species of China (Fang et al. Citation1999), which is known for their commercially and ecologically forest trees (Hanzeh and Dayanandan Citation2004). It mainly occurs in the area of Gansu, Shaanxi, Sichuan, Guizhou, and Yunnan province. However, it is often confused with P. davidiana, and the systematic of the species is unclear (Fang et al. Citation1999). For a better understanding of the relationships of P. rotundifolia var. duclouxiana and P. davidiana, it is necessary to reconstruct a phylogenetic tree of the Populus species based on high-throughput sequencing approaches. Mitochondrial (mt) genome has drawn increased attention during the post genomic eras owing to its maternal pattern of inheritance and unique evolutionary features, and the genetic information contained within the mt genome has been an important source of information for resolving relationships among plants (Gualberto et al. Citation2014; Dames et al. Citation2015; Satjarak et al. Citation2017). Therefore, we sequenced and assembled the complete mt genome of P. rotundifolia var. duclouxiana, and this information might provide further understanding of the origin and genetic differentiation with other Populus species.
The fresh leaves of P. rotundifolia var. duclouxiana were sampled from Kunming (Yunnan, China; Coordinates: 102°45′40″E, 25°3′42″N; altitude: 1940 m). Total genome DNA was extracted with the Ezup plant genomic DNA preps Kit (Sangon Biotech, Shanghai, China). All the voucher specimens of P. rotundifolia var. duclouxiana were preserved in the Herbarium of Southwest Forestry University (contact Dan Zong, [email protected]) under the voucher number (SWFU-QXY-01), and DNA samples were stored at −80 °C at the Key Laboratory of State Forestry Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, China. Total DNA was used to generate libraries with an average insert size of 400 bp and sequences using the Illumina HiseqX platform. Approximately 15.0 GB of raw data was generated with 150 bp paired-end read lengths. Then, the raw data were used to assemble the complete mt genome using the software of GetOrganelle (Jin et al. Citation2020) with P. davidiana as the reference (Choi et al. Citation2017). Genome annotation was performed with the program Geneious R8 (Biomatters Ltd., Auckland, New Zealand), by comparing with the mt genome of P. davidiana. The mtDNA sequence with complete annotation information was deposited at GenBank database under the accession number MW566588.
The complete mt genome of P. rotundifolia var. duclouxiana is a typical circular molecular of 772,549 bp in length. The overall GC content is 44.8%, with a base composition of 27.5% A, 22.4% C, 22.4% G, and 27.6% T. The P. rotundifolia var. duclouxiana mt genome contains 58 typical mt genes, including 33 protein-coding genes, 22 tRNA genes, and three rRNA genes (rrn5, rrnL, and rrnS). Eight genes contain introns, gene ccmFc, rpl2, and rps3 were interrupted by a single intron, gene nad4 contained three introns and gene nad1, nad2, nad5, and nad7 have four introns. Gene nad1, nad2, and nad5 are trans-splicings.
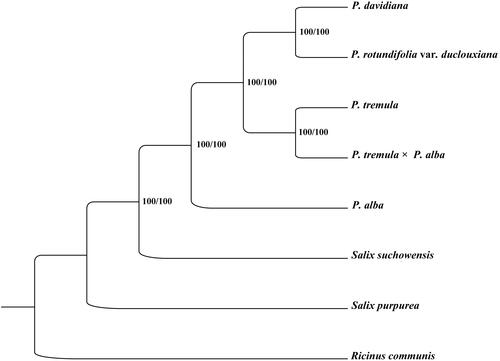
To determine the phylogenetic location of P. rotundifolia var. duclouxiana with respect to the other Salicaceae with fully sequenced mt genomes, the complete P. rotundifolia var. duclouxiana mt genome was used to reconstruct the phylogenetic relationships. Seven complete mt genomes of Salicaceae were achieved from GenBank, and the species of Ricinus communis in the family of Euphorbiaceae was used as outgroup. All of these eight complete mt sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). A maximum-likelihood (ML) tree was inferred by bootstrapping with 1000 replicates through IQ-TREE 1.5.5 (Nguyen et al. Citation2015) based on the TPM3u + F nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). The phylogenetic analysis showed that P. rotundifolia var. duclouxiana appeared to be closely related to P. davidiana ().
Figure 1. Phylogenetic relationships among seven complete mitochondrial genomes of Salicaceae. Bootstrap support values are given at the nodes. Mitochondrial genome accession number used in this phylogeny analysis: P. davidiana: KY216145; P. rotundifolia var. duclouxiana: MW566588; P. tremula: KT337313; P. tremula×alba: KT429213; P. alba: MK034705; Salix suchowensis: KU056812; Salix purpurea: KU198635; Ricinus communis: HQ874649.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW566588. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA748199, SAMN20309755, and SRR15195673, respectively.
Additional information
Funding
References
- Choi MN, Han M, Lee H, Park HS, Kim MY, Kim JS, Na YJ, Sim SW, Park EJ. 2017. The complete mitochondrial genome sequence of Populus davidiana Dode. Mitochondrial DNA Part B. 2(1):113–114.
- Dames S, Eilbeck K, Mao R. 2015. A high-throughput next-generation sequencing assay for the mitochondrial genome. Methods Mol Biol. 1264:77–88.
- Fang ZF, Zhao SD, Skvortsov AK. 1999. Flora of China (English version). Vol. 4. Beijing: Science Press; p. 139–274.
- Gualberto JM, Mileshina D, Wallet C, Niazi AK, Weber-Lotfi F, Dietrich A. 2014. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 100(1):107–120.
- Hanzeh M, Dayanandan S. 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am J Bot. 91:1398–1408.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assemble of organelle genomes. Genome Biol. 21:241.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Satjarak A, Burns JA, Kim E, Graham LE. 2017. Complete mitochondrial genomes of prasinophyte algae Pyramimonas parkeae and Cymbomonas tetramitiformis. J Phycol. 53(3):601–615.
