Abstract
The peach (Prunus persica L. Batsch) is one of the important stone fruit crops in the Crimea Peninsula and the southern part of Russia. The complete chloroplast genome of the peach cultivar ‘Sovetskiy’ is published in this paper. The chloroplast genome size is 157,756 bp. It contains 126 genes, including 81 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. The chloroplast genome also contains a large single-copy region of 85,960 bp, a small single-copy (SSC) region of 19,045 bp, and two inverted repeats regions of 26,375 bp and 26,372 bp. The overall base composition of the genome in descending order is 31.2% – A, 32.1% – T, 18.7% – C, and 18.0% – G. The total GC content of the chloroplast genome is 36.7%. Maximum-likelihood phylogenetic analysis involving nine chloroplast genomes of the Prunus genus revealed a separate cluster for P. persica and its possible landrace – P. ferganensis.
The peach (Prunus persica L. Batsch) is a species of the Rosaceae family (Bielenberg et al. Citation2009). This perennial stone fruit tree was domesticated at least 7500 years ago in China (Zheng et al. Citation2014). Peach tree is widely cultivated worldwide in continental or temperate climates (Li et al. Citation2019). The natural conditions of the Crimea Peninsula are favorable for peach horticulture development. Nikita Botanical Garden has one of the largest peach germplasm collections which contain more than 900 cultivars and forms. Moreover, new cultivars with important agricultural traits are actively created by the local breeders (Smykov Citation2019). In this paper, we present a complete assembly of the chloroplast genome of the peach cultivar Prunus persica cv. ‘Sovetskiy’ (Yezhov et al. Citation2005), which is known as one of the first cultivars created in the Soviet Union by crossing the ‘Golden Jubilee’ and ‘Narindzhi Pozdny’ cultivars. The peach cultivar ‘Sovetskiy’ was bred by Ivan Ryabov in the early 1950s and belongs to the Iranian peach cultivar group (Yezhov et al. Citation2005).
Genomic DNA was extracted from the ‘Sovetskiy’ cultivar (Nikita Botanical Garden, collection number − 40) leaves following the modified methodology described previously (Lo Piccolo et al. Citation2012). The Rapid Sequencing Kit (SQK-RAD004) and GridION genome analyzer (Oxford Nanopore Technologies, Kidlington, UK) were used for the genomic DNA sequencing.
535,545 GridION long reads (average length: 34.16 kbp) were generated for ‘Sovetskiy’ cultivar peach specimen. Obtained reads were mapped to the P. persica chloroplast reference genome (NC_014697.1) using minimap2 (Li Citation2018) with default parameters (coverage was 2185 X). Only 979,641 chloroplast genome reads were used for subsequent de novo assembly.
De novo assembly of the P. persica cv. ‘Sovetskiy’ chloroplast genome was performed using the Flye program (Kolmogorov et al. Citation2019) with polisher iterations. The chloroplast genome was annotated by using the CHloe web service (https://chloe.plantenergy.edu.au/).
The chloroplast genome of P. persica cv. ‘Sovetskiy’ (GenBank MZ065355) is 157,756 bp in length and has a circular structure. Chloroplast genome contains 126 genes, including − 81 protein-coding gene (PCG), eight ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. Among these genes, 16 genes (ndhB, petB, petD, rpl2, trnA-UGC, trnG-UCC, trnI-GAU, trnL-UAA, atpF, ndhA, rpl16, rpoC1, rps12A, rps16, trnK-UUU, trnV-UAC) have one intron and three genes (ycf3, rps12B, clpP1) have two introns. In this genome, eight PCG (ndhB, rpl2, rpl23, rps12B, rps19, rps7, ycf1, and ycf2), four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) are duplicated, the others genes have a single copy.
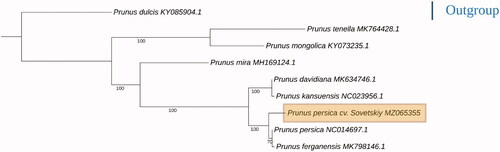
The complete chloroplast genome sequences of eight species of the Prunus genus were aligned with each other using the MUSCLE v3.8.1551 program (Edgar Citation2004) and then were used to construct phylogenic maximum-likelihood (ML) tree using the RaxML software v8.0.0 (Stamatakis et al. Citation2008) with 1000 bootstrap parameter. The complete chloroplast genome of almond (P. amygdalus) (KY085904.1) was used as an outgroup. Phylogenetic analysis based on the whole chloroplast sequences of Prunus species presents ‘traditional’ topology () which has been previously described. Cultivar ‘Sovetskiy’ was clustered with the cultivar ‘Nemared’ (NC014697.1) and Fergana peach – P. ferganensis (MK798146.1) which is usually considered as a landrace of P. persica (Verde et al. Citation2013; Xin et al. Citation2021).
This study represents the first chloroplast genome assembly and annotation of cultivated P. persica cv. ‘Sovetskiy’. These data will be a potential genetic resource for peach cultivar identification and breeding.
Acknowledgements
This work was done in the Kurchatov Center for Genome Research and in the Kurchatov Genomic Center – NBG-NSC. This work was carried out using high-performance computing resources of federal center for collective usage at NRC ‘Kurchatov Institute’, http://computing.kiae.ru/.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
A specimen is grown at Nikita Botanical Garden (http://nbgnsc.ru/en, contact person: Anatoliy Smykov – e-mail: [email protected]) under the voucher number 40. A genomic DNA is deposited at National Research Center ‘Kurchatov Institute’ (http://eng.nrcki.ru/, contact person: Svetlana Tsygankova – e-mail: [email protected]) under the voucher number PP1C. The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MZ065355. The associated BioProject, SRA, and sample numbers are PRJNA742656, SRR15000149, and SAMN19967399, respectively.
Additional information
Funding
References
- Bielenberg D, Gasic K, Chaparro JX. 2009. An introduction to peach (Prunus persica). Vol. 6. New York: Springer.
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5:113.
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100.
- Li Y, Cao K, Zhu G, Fang W, Chen C, Wang X, Zhao P, Guo J, Ding T, Guan L, et al. 2019. Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol. 20(1):36.
- Lo Piccolo S, Alfonzo A, Conigliaro G, Moschetti G, Burruano S, Barone A. 2012. A simple and rapid DNA extraction method from leaves of grapevine suitable for polymerase chain reaction analysis. Afr J Biotechnol. 45(11):10305–10309.
- Smykov AV. 2019. Condition and perspectives of horticulture development in the south of Russia. Acta Hortic. 1255(1255):1–6.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771.
- Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F. 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 45(5):487–494.
- Xin L, Yu Z, Jiying G, Jianbo Z, Quan J, Fei R. 2021. The complete chloroplast genome sequence of cultivated peach (Prunus persica var. nectarina cv. 'Rui Guang 18'). Mitochondrial DNA B Resour. 6(1):208–210.
- Yezhov VN, Smykov AV, Smykov VK, Khokhlov SY, Zaurov DE, Mehlenbacher SA, Molnar TJ, Goffreda JC, Funk CR. 2005. Genetic resources of temperate and subtropical fruit and nut species at the Nikita Botanical Gardens. HortScience. 40(1):5–9.
- Zheng Y, Crawford GW, Chen X. 2014. Archaeological evidence for peach (Prunus persica) cultivation and domestication in China. PLOS One. 9(9):e106595.