Abstract
Dudusa sphingiformis is an important lepidopteran pest widely distributed in tropical and subtropical zones of Asia. In this paper, the complete mitochondrial genome (mitogenome) of D. sphingiformis was determined by next-generation sequencing. The mitogenome was 15,806 bp in length, comprising 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and an AT-rich control region (D-loop). The gene arrangement of this mitogenome was identical to that of the previous studies of Notodontidae moths. Almost all the PCGs initiated with typical ATN codons, except for cox1 with CGA. Among them, nine PCGs terminated with TAA or TAG, while other four PCGs (cox1, cox2, nad5, and nad4) with incomplete stop codon T. All the 22 tRNAs had the typical cloverleaf structure, except for trnS1, whose dihydrouridine (DHU) arm forms a simple loop. Phylogenetic analysis based on the concatenated nucleotide sequences of 13 PCGs indicated that D. sphingiformis was more closely related to other species of family Notodontidae, forming a monophyletic group, with well-resolved relationships among five family of Noctuoidea.
Notodontidae consists of approximately 3800 known species, is a group of moths belong to superfamily Noctuoidea, which is the biggest superfamily of Lepidoptera (Miller Citation1992). Almost all of species Notodontidae are important injurious insects for forestry and farming (Miller Citation1991). To date, there have been several studies focused on the phylogenetic analysis of Notodontidae using molecular markers (Regier et al. Citation2017), which significantly promoted the systematic studies of Lepidoptera. The mitochondrial genome (mitogenome) provides important information for phylogenetic analysis and studies of evolutionary history (Cameron Citation2014). However, the complete mitogenome of the majority of species in family Notodontidae remain unsolved, expect for two species: Clostera anachoreta (Zhu et al. Citation2017) and Clostera anastomosis (Zhu et al. Citation2018). Dudusa sphingiformis Moore, 1872 is a member of the family Notodontidae, and widely distributed in tropical and subtropical zones of Asia (Jung and Oh Citation2012). This moth feeds on branches and leaves of deciduous tree, and is an important pest for agricultural industry. In the present study, we first sequenced, assembled, and annotated the complete mitochondrial genome of D. sphingiformis and further reconstructed the phylogenetic relationships combining with available mitogenomes of other species of Noctuoidea in GenBank.
Specimens of D. sphingiformis were collected in June 2020 in Huixian County of Gansu province, China (33°39′35.7″N; 106°16′11.5″E), and stored in the Institute of Zoology and Ecology, College of Life Science, Northwest Normal University, Lanzhou, China (accession number: LN2020013). The genomic DNA was extracted from muscle tissue of a single specimen’s thorax, which was sequenced by Illumina NovaSeq 6000 platform with both directions of 150 bp reads. The MitoZ v2.3 (Meng et al. Citation2019) was used to assemble the mitogenome based on 6 Gb clean data. The assembled mitogenome was annotated using the MITOS web server (Bernt et al. Citation2013) under the invertebrate mitochondrial code. The tRNA genes were confirmed by ARWEN online application (Laslett and Canback Citation2008). The ClustalX 2.0 software (Larkin et al. Citation2007) was used to align sequence dataset for phylogenetic analysis. The phylogenetic tree was built using W-IQ-TREE (Trifinopoulos et al. Citation2016). The newly determined genome from the present study was deposited in GenBank database (accession number: MW788876.1).
The complete mitogenome of D. sphingiformis was 15,806 bp in length. The A + T content of the whole genome sequence was 81.2% (40.7% A, 40.5% T, 11.7% C, and 7.1% G), indicating significant A + T bias. This mitogenome contained 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and AT-rich control region (D-loop). All of genes shared identical pattern of gene arrangement with other species of Notodontidae (Zhu et al. Citation2017, Citation2018). Like other moths of Noctuoidea (Cao et al. Citation2012), almost all the PCGs began with typical ATN codons (seven ATG, one ATA, and four ATT), except for cox1 with CGA. Among them, nine PCGs terminated with TAA or TAG, while other four PCGs (cox1, cox2, nad5, and nad4) with incomplete stop codon T. All the 22 tRNAs, ranging from 65 to 72 bp, had the typical cloverleaf structure, except for trnS1, whose dihydrouridine (DHU) arm formed a simple loop. The absence of the DHU arm in trnS1 was found in the mitochondrial genomes existed in most insects (Wolstenholme Citation1992). The two ribosomal RNA genes, rrnL and rrnS, were 1379 bp and 838 bp in length, respectively. The control regions was 297 bp long with 93.6% A + T content. This condensation or conciseness of control region was commonly demonstrated by other species of superfamily Noctuoidea (e.g. accession number: KX108766.1 and MH286069.1).
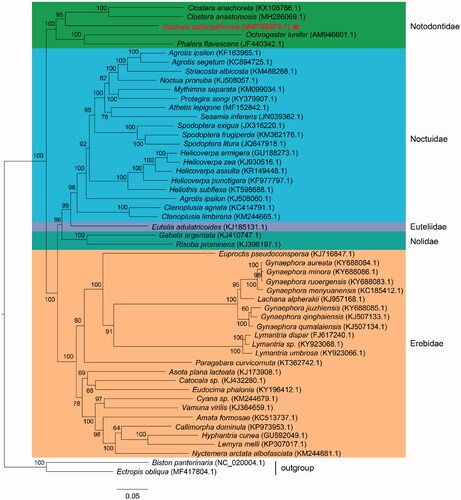
Until now, there are five species of Notodontidae, whose mitochondrial genomes with annotation have been reported. We reconstructed the phylogenetic relationships of superfamily Noctuoidea combining with available mitogenomes of other species of Noctuoidea in GenBank. These species including D. sphingiformis in this study belong to five family: Notodontidae, Noctuidae, Euteliidae, Nolidae, and Erebidae. We performed a maximum-likelihood (ML) analysis based on the best-fitting substitution model of GTR + F+I + G4 according to BIC with 1000 ultrafast bootstrap replicates. The concatenated nucleotide sequences of 13 PCGs from 49 species of superfamily Noctuoidea were used to construct the phylogenetic tree. Two species of family Geometridae: Ectropis oblique (accession number: MF417804.1) and Biston panterinaria (accession number: NC_020004.1) were used as outgroup for phylogenetic analyses. The results indicated that D. sphingiformis showed the closest phylogenetic relationship with other species of family Notodontidae, which clustered into a monophyletic group (). The topology structure demonstrated a clear phylogenetic relationship of family Notodontidae with other populations of superfamily Noctuoidea, which were supported by the previous phylogenetic studies of Noctuoidea (Zhu et al. Citation2018). In this study, we characterized the complete mitochondrial genome of D. sphingiformis and provided new additions to studies of phylogeny construction related to Notodontidae species and other moths.
Figure 1. Maximum-likelihood tree showing phylogenetic relationships of Dudusa sphingiformis and other 49 species of superfamily Noctuoidea (including two species of superfamily Geometridae as outgroup) based on GTR + F+I + G4 model, using concatenated nucleotide sequences of 13 protein-coding genes. Numbers above or below nodes indicated the ultrafast bootstrap support values estimated with 1000 replicates. The family names and outgroup related to phylogenetic analysis were depicted at right side. The five family included Notodontidae, Noctuidae, Euteliidae, Nolidae, and Erebidae. The newly sequenced mitogenome was highlighted by the star.
Acknowledgements
We would like to thank Yimin Du, Hui Zhang, and Fan Zhao for their experimental assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov under the accession number MW788876.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715946, SRR14018638, and SAMN18388692, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13(1):276.
- Jung SH, Oh HS. 2012. Insecta (Lepidoptera) of Yeongsil in Hallasan Mountain National Park. J Korean Nat. 5(2):181–192.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Miller JS. 1991. Cladistics and classification of the Notodontidae (Lepidoptera: Noctuoidea) based on larval and adult morphology. B Am Mus Nat Hist. 240(204):1–230.
- Miller JS. 1992. Host–plant associations among prominent moths: lineages within the moth family Notodontidae show contrasting host-use patterns. BioScience. 42(1):50–57.
- Regier JC, Mitter C, Mitter K, Cummings MP, Bazinet AL, Hallwachs W, Janzen DH, Zwick A. 2017. Further progress on the phylogeny of Noctuoidea (Insecta: Lepidoptera) using an expanded gene sample. Syst Entomol. 42(1):82–93.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wolstenholme DR. 1992. Genetic novelties in mitochondrial genomes of multicellular animals. Curr Opin Genet Dev. 2(6):918–925.
- Zhu XY, Xin ZZ, Liu Y, Wang Y, Huang Y, Yang ZH, Chu XH, Zhang DZ, Zhang HB, Zhou CL, et al. 2018. The complete mitochondrial genome of Clostera anastomosis (Lepidoptera: Notodontidae) and implication for the phylogenetic relationships of Noctuoidea species. Int J Biol Macromol. 118(Pt B):1574–1583.
- Zhu XY, Xin ZZ, Wang Y, Zhang HB, Zhang DZ, Wang ZF, Zhou CL, Tang BP, Liu QN. 2017. The complete mitochondrial genome of Clostera anachoreta (Lepidoptera: Notodontidae) and phylogenetic implications for Noctuoidea species. Genomics. 109(3–4):221–226.
