Abstract
Schnabelia tetrodonta is a medicinal plant used in traditional Chinese medicine. However, the molecular biology data of the species was too scarce to bioprospect the medicinal species. In this study, the first complete chloroplast genome (cp) of S. tetrodonta was sequenced and assembled based on the next generation sequencing. The cp genome is 157,004 bp in length, including a large single-copy (LSC) region of 83,605 bp, a small single-copy (SSC) region of 36,899 bp, and a pair of inverted repeat (IR) regions of 18,250 bp each. The genome encodes 134 genes, including 90 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The GC content of whole genome is 37.80%. The phylogenetic analysis based on 20 complete cp sequences (19 genome sequences from the Teucrioideae of Lamiaceae and an outgroup of Ipomoea purpurea) revealed that S. tetrodonta was closely related to S. oligophylla.
Schnabelia tetrodonta (Sun) C. Y. Wu et C. Chen is a member of the genus Schnabelia in Lamiaceae. It is an Endangered species, narrowly distributed in the mountainous areas of central Sichuan province and northern Guizhou province of China (Li and Hedge Citation1994). Schnabelia tetrodonta is rich in flavonoids, polysaccharides and saponins (Dou et al. Citation2002, Citation2003; Zhou et al. Citation2004). It is used as a traditional Chinese medicine to treat rheumatic joint pain. Modern pharmacological studies show that S. tetrodonta has anti-fatigue, anti-oxidant, anti-inflammatory, analgesic and immunomodulatory effects (Ouyan). However, S. tetrodonta, as an important medicinal plant, has no genome information reported up to now. In this study, the first chloroplast (cp) genome of the species was sequenced for excavating and protecting the resource of S. tetrodonta.
Fresh leaves of S. tetrodonta were collected from Nanchuan, Chongqing, China (107°21′ E, 29°13′ N, 591 m). The voucher specimen was conserved in Chongqing Institute of Medicinal Plant Cultivation under the accession number of CIMPC-RFM-20210301 (Contact person: Fengming Ren; Email: [email protected]). A modified CTAB-based method was used to extract the whole genomic DNA, and the purity and integrity of the DNA were analyzed by Nanodrop (Thermo Fisher Scientific) and agarose gel electrophoresis. Total DNA was used to generate libraries with insert size of 350 bp and was generated 3.43 Gb raw reads by Illumina Hiseq 2500 Platform (Illumina, Hayward, CA, USA). Low-quality readings and adapters in the raw data were deleted by trimmomati (version 0.35) with default paprameters (Bolger et al. Citation2014). Using the clean data with 150 bp paired-end read lengths obtained from the raw data, a cpgenome was assembled by NOVOPlasty (version 4.1) with the default parameters (Nicolas et al. Citation2016) and annotated by CPGAVAS2 (Shi et al. Citation2019). After manual check and adjustment, the annotated cp genome was submitted to GenBank (MW928532). The complete cp genome of S. tetrodonta was 157,004 bp in size and exhibited a typical angiosperm circular cp structure, containing four regions: large single-copy region (LSC: 96,530 bp), small single-copy region (SSC: 9,636 bp), and a pair of inverted repeats (IR: 41,955 bp). The GC content was 37.80% (whole genome), 36.16% (LSC), 37.72% (SSC) and 41.64% (SSC). The GC content of the genome and each genomic region was also typical of angiosperm cp structure. The genome encoded 134 genes, including 90 protein-coding genes, 36 tRNA genes, and 8 rRNA genes.
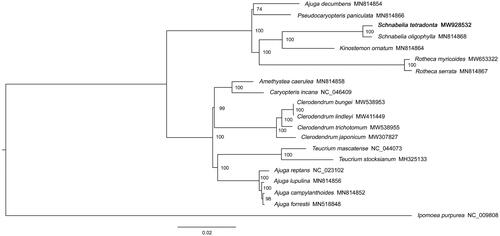
Nineteen genome sequences from the Teucrioideae of Lamiaceae were downloaded from the NCBI database. The genome sequence of Ipomoea purpurea Lam. was used as an outgroup. Finally, a total of twenty cp genome sequences were multi-aligned by MAFFT software (version 7.487) with the parameter of ‘–auto’ (Katoh and Standley Citation2013). Based on the aligned sequences, a Maximum-likelihood phylogenetic tree was built by IQ-TREE (version 1.6.12) (Lam-Tung et al. Citation2015) with 1000 bootstrap replicates under parameters of ‘-nt AUTO -m MFP -bb 1000 -bnni’. Phylogenetic analysis showed that S. tetrodonta was closely related to S. oligophylla (). Schnabelia oligophylla is the only other species of Schnabelia with available plastome data. So the result is expected, which also support the reliability of our data. However, the reliability of the phylogenetic state of S. tetrodonta was not high because of scarce available Schnabelia plastome data. To eliminate the phylogenetic state of Schnabelia, more available Schnabelia plastome data should be produced in the further studies. In this study, to sequence the plastome of S. tetrodonta is a step forward in tackling this lack of information.
Figure 1. Maximum likelihood phylogenetic tree based on the chloroplast genome sequences of 19 Teucrioideae (Lamiaceae) species and Ipomoea purpurea (outgroup). The GenBank accession numbers is behind the Latin name. The bootstrap support values are beyond each node in the tree S. tetrodonta is marked by bold font.
Acknowledgements
We thank Professor Qiuping Tan for providing plant material.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession No. MW928532. The associated Bio-Project, Bio-Sample and SRA numbers are PRJNA543381, SAMN19416297, and SRR14688233, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dou H, Liao X, Peng SL, Pan YJ, Ding LS. 2003. Chemical constituents from the roots of Schnabelia tetradonta. HCA. 86(8):2797–2804.
- Dou H, Zhou Y, Chen C, Peng S, Liao X, Ding L. 2002. Chemical constituents of the aerial parts of Schnabelia tetradonta. J Nat Prod. 65(12):1777–1781.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lam-Tung N, Schmidt HA, Arndt VH, Bui QM. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li HW, Hedge IC. 1994. Lamiaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 17. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 250–299.
- Nicolas D, Patrick M, Guillaume S. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. (4): 1–9.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated n plastome sequence annotator and analyzer. Nucleic Acids Res. 47(1):65–73.
- Zhou Y, Dou H, Peng S, Zhang X, Ding L. 2004. Structural characterization of aminoethylphenyl oligoglycosides in Schnabelia tetradonta. Rapid Commun Mass Spectrom. 18(10):1172–1176.
