Abstract
A non-biting midge Chironomus yoshimatsui has been widely used in ecotoxicology and chemical risk assessments. In this study, the complete mitochondrial genome (mitogenome) sequence of C. yoshimatsui was determined using short-read next-generation sequencing technologies. The mitogenome was 15,734 bp in length and consisted of 13 protein coding genes, 2 ribosomal RNAs, and 22 transfer RNAs. The A + T content was 77.8%. The gene order was identical to the pattern conserved across Diptera. The mitocgenome sequence obtained in this study provides a useful resource for further evolutionary and ecological studies.
The non-biting midge Chironomus yoshimatsui Martinet Sublette, 1972 is widely distributed in freshwater environments in Japan and Korea, and has been used in ecotoxicological studies and chemical risk assessments (Sugaya Citation1997; OECD Citation2004; Ishimota et al. Citation2020). Despite its ecological and ecotoxicological value, to date, the sequence information of this species is available for limited fragments, such as partial cytochrome c oxidase subunit I (cox1) (Kondo et al. Citation2016; Kawai et al. Citation2019) and heat shock protein 70 (Yoshimi et al. Citation2002). In this study, we determined the complete mitochondrial genome (mitogenome) sequence of C. yoshimatsui, and performed a phylogenetic analysis among Chironomidae family with the publicly available data.
The specimen of Chironomus yoshimatsui was obtained from laboratory culture at the National Institute for Environmental Studies, Japan. The laboratory culture originated from a population that was collected from a drain ditch at Yumoto, Nikko, Tochigi (36°48'21"N, 139°25'26"E) in 1991 and 1993 (Sugaya Citation1997) and morphologically identified as C. yoshimatsui according to Sasa (Citation1978). DNA was extracted from whole eggs using DNeasy Blood & Tissue Kit (Qiagen) and sequenced on an Illumina MiSeq with a paired-end library. The extracted DNA was deposited in the Ecotoxicity Research Section at the National Institute for Environmental Studies, Japan (NIES-202106-MID1, Kyoshiro Hiki, [email protected]). The sequence data (INSDC accession number: DRR304759) were trimmed and assembled using CLC Genomics Workbench (ver. 21.0.4). The raw reads were trimmed with a Phred score <20 after removal of adapter sequences and reads containing more than five ambiguous nucleotides. Reads shorter than 50 bases were discarded. The remaining reads were de novo assembled with the default parameters. The assembled mitogenome was annotated by MITOS 2 webserver (Bernt et al. Citation2013) and by manual comparison with orthologous genes of other midge species. Phylogenetic analyses were performed based on 13 protein coding gene (PCG) and 2 ribosomal RNA (rRNA) sequences in the mitogenome of C. yoshimatsui and those of other Chironomidae species. Each sequence was aligned separately using MAFFT (ver. 7.453) with the L-INS-i option (Katoh and Standley Citation2013) and then the alignments were filtered using trimAl (ver. 1.4.1) (Capella-Gutiérrez et al. Citation2009) with the heuristic method. Maximum likelihood-based phylogenetic trees were inferred using IQ-TREE (ver. 1.6.12) (Nguyen et al. Citation2015) with the ‘–spp’option to allow partition-specific evolution rates and visualized by FigTree (ver. 1.4.4). The best-fit model for each PCG was determined by ModelFinder (Kalyaanamoorthy et al. Citation2017) implemented in IQ-TREE, based on Akaike information criteria.
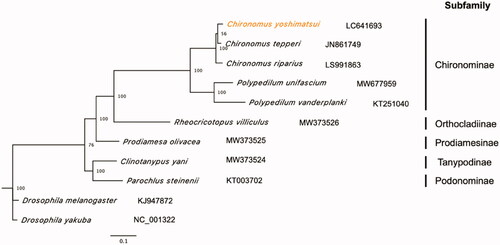
The complete circular mitogenome of 15,734 bp was obtained for C. yoshimatsui (INSDC accession number: LC641693) with 27× mean coverage. The mitogenome contained typical gene components, including 13 PCGs, 2 rRNAs, and 22 transfer RNAs (tRNAs). The order of 37 genes was completely identical to the pattern conserved across Diptera species such as Drosophila yakuba and D. melanogaster (Boore Citation1999). Among 37 genes, four PCGs (nd1, nd4, nd4l, and nd5), eight tRNAs (trnC, trnF, trnH, trnL [UAG], trnP, trnQ, trnV, and trnY), and two rRNAs located on the light strand, and the other 23 genes located on the heavy strand, which is consistent with the mitogenomes of other Chironomidae species (Zheng et al. Citation2021). The ratios of A + T and G + C nucleotides of 13 PCGs were 75.1 and 24.9%, respectively, while those ratios of the entire sequences were 77.8 and 22.2%, respectively. All 13 PCGs initiated with the standard start codons (i.e. ATT, ATA, and ATG) and had complete stop codons (i.e. TAA). The cox1 sequence was completely identical to the previously reported partial sequence of cox1 of this species (accession number: AB740260) collected at Ibaraki, Japan (Kondo et al. Citation2016). The maximum likelihood-based phylogenetic analysis showed that C. yoshimatsui was grouped with the other two Chironomus species with high bootstrap values () and these three species were grouped with other two species belonging to the subfamily Chironominae (Polypedilum unifascium and Polypedilum vanderplanki) in one clade within the Chironomidae family.
Figure 1. Maximum likelihood tree based on 13 PCGs and 2 rRNAs in mitochondrial genomes of Chironomidae family. Orange represents the genome obtained in this study. Drosophila melanogaster and D. yakuba were used as the outgroup. Non-parametric bootstrap values (based on 3,000 times resampling) are shown at nodes. The scale bar indicates the number of amino acid substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data that support the findings of this study are openly available at INSDC nucleotide database with the accession number LC641693, and at Sequence Read Archives with the accession number PRJDB11895 (BioProject), SAMD00387595 (BioSample), DRX294195 (Experiment), and DRR304759 (Run).
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Ishimota M, Tajiki-Nishino R, Fukuyama T, Tomiyama N. 2020. Rapid adaptation of Chironomus yoshimatsui to acetylcholinesterase inhibitors (pyraclofos and pirimicarb) in a multi-generation study. J Environ Sci Heal Part B Pestic Food Contam Agric Wastes. 55(5):429–437.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kawai K, Kawaguchi K, Kodama A, Saito H. 2019. Fundamental studies on acid-tolerant chironomids in Japan. Limnology. 20(1):101–107.
- Kondo NI, Ueno R, Ohbayashi K, Golygina VV, Takamura K. 2016. DNA barcoding supports reclassification of Japanese Chironomus species (Diptera: Chironomidae). Entomol. Sci. 19(4):337–350.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- OECD. 2004. Test No. 218: sediment-water chironomid toxicity using spiked sediment, OECD guidelines for the testing of chemicals, section 2. OECD. https://doi.org/10.1787/9789264070264-en.
- Sasa M. 1978. A comparative study of adults and immature stages of nine Japanese species of the genus Chironomus (Diptera, Chironomidae). Res. Rep. from Natl. Inst. Environ. Stud. 3:1–63.
- Sugaya Y. 1997. Intra-specific variations of the susceptibility to insecticides in Chironomus yoshimatsui. Med Entomol Zool. 48(4):345–350.
- Yoshimi T, Minowa K, Karouna-Renier NK, Watanabe C, Sugaya Y, Miura T. 2002. Activation of a stress-induced gene by insecticides in the midge, chironomus yoshimatsui. J Biochem Mol Toxicol. 16(1):10–17.
- Zheng CG, Zhu XX, Yan LP, Yao Y, Bu WJ, Wang XH, Lin XL. 2021. First complete mitogenomes of diamesinae, orthocladiinae, prodiamesinae, tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ. 9:e11294.
