Abstract
Prunus clarofolia is an endemic species that widely distributed in subtropical regions of China. Here we present its complete plastome. The plastome of P. clarofolia is successfully assembled from raw reads sequenced by Illumina Hiseq 2500 platform system. The complete chloroplast of this species is 158,053 bp in length with 36.7% overall GC content, including a pair of invert repeat regions (IR) (26,393bp) that is divided by a large single copy region (LSC) (86,088bp) and a small single copy region (SSC) (19,179bp). The plastid genome contained a total of 130 genes, including 85 coding genes, 8 rRNA genes, and 37 tRNA genes. Each of rps16, atpF, rpoC1, clpP, petB, petD, rpl16, rpl2, ndhB, and ndhA contains one intron, rps12 and ycf3 contains two introns. Phylogenetic analysis indicates that P. clarofolia has a closer relationship with P. avium.
Keywords:
Prunus L. subg. Cerasus (Mill.) A. Gray contains almost 150 species that mainly distribute in temperate and subtropical regions of the northern hemisphere (Yu et al. Citation1986). Recent studies have shown that frequent interspecific hybridizations and non-significant morphological differences have complicated the taxonomy of this subgenus (Ohta et al. Citation2007; Shi et al. Citation2013; Zhang et al. Citation2017). Prunus clarofolia C.K. Schneid (Fedde Citation1905) wildly spread in subtropical regions of China, and its leaves and bracts with sparse pubescent and umbellate inflorescences are often regarded as unique features. However, there still remains blank about the genetic relationship between this important wild species which are distributing in China and other subg. Cerasus species. Therefore, we sequenced the whole chloroplast genome of P. clarofolia to elucidate its phylogenetic relationship with other subg. Cerasus species.
The plant material was obtained from Songyang, Zhejiang province, China (28°87′56″N 119°28′35″E, altitude 357 m). A specimen was deposited at Nanjing Forestry University (https://shengwu.njfu.edu.cn/; collector: Meng Li, [email protected]; voucher number: NF: 161098736). Total DNA was extracted from fresh leaves with a modified CTAB protocol (Cai et al. Citation2014). The whole genome sequencing was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA). A total of 1.35 Gb of clean paired-end reads (Phred scores >20) were assembled using the programme GetOrganelle v1.7.5 (Jin et al. Citation2020). The plastome was annotated by the web application GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017).
The total length of complete chloroplast genome of P. clarofolia is 158,053 bp, with a total GC content of 36.7%. The plastid genome has a typical quadripartite structure, including large single-copy region (LSC) of 86,088bp, small single-copy region (SSC) of 19,179 bp, and a pair of inverted repeat regions (IRA and IRB) of 26,393bp each. The complete cp genome contains 130 genes, including 85 protein-coding genes, 37 tRNA, and. 8 rRNA genes. In this plastome genome, rps16, atpF, rpoC1, clpP, petB, petD, rpl16, rpl2, ndhB and ndhA contains an intron while rps12 and ycf3 contains 2 introns.
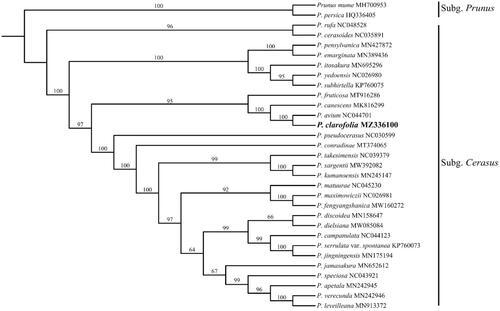
To determine the phylogenetic position of P. clarofolia, the complete chloroplast genome of P. clarofolia was aligned with other 28 subg. Cerasus species and 2 outgroups of subg. Prunus from GenBank using MAFFT v7.467 (Katoh et al. Citation2005) and visually checked and adjusted in Bioedit. Maximum-likelihood (ML) analysis was conducted in IQ-TREE v2.1.1 (Vergara et al. Citation2015) with 1000 bootstrap replications. The result was well-resolved and revealed that P. clarofolia was belonged to subg. Cerasus and was most closely related to P. avium (). In summary, the complete plastid genome of P. sargentii will provide useful genetic information for increasing the richness of subg. Cerasus, as well as assisting in phylogenetic and evolutionary studies of subg. Cerasus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The plastid genome in this study is available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with an accession number MZ336100, and SRA submitted to NCBI under the BioProject No. PRJNA732592, Biosample NO. SAMN19324472, and SRA number: SRR14661355.
Additional information
Funding
References
- Cai WJ, Xu DB, Lan X, Xie HH, Wei JG. 2014. A new method for the extraction of fungal genomic DNA. Agric Res Appl. 152:1–5.
- Fedde F. 1905. Repertorium specierum novarum regni vegetabilis. Vol. 1. Berlin, Germany; p. 67.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Ohta S, Yamamoto T, Nishitani C, Katsuki T, Iketani H, Omura M. 2007. Phylogenetic relationships among Japanese flowering cherries (Prunus subgenus Cerasus) based on nucleotide sequences of chloroplast DNA. Plant Syst Evol. 263(3–4):209–225.
- Shi S, Li JL, Sun JH, Yu J, Zhou SL. 2013. Phylogeny and Classification of Prunus sensu lato (Rosaceae). J Integr Plant Biol. 55(11):1069–1079.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Vergara D, White K, Keepers K, Kane N. 2015. The complete chloroplast genomes of Cannabis sativa and Humulus lupulus. Mitochondrial DNA. 27:1–2.
- Yu TT, Lu LT, Ku TC, Li CL, Chen SX. 1986. Rosaceae (3) Prunoideae. In: Yu TT, editor. Flora Reipublicae Popularis Sinicae, Tomus 38. Beijing: Science Press.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 214(3):1355–1367.