Abstract
Chrysosplenium nudicaule Bunge, Tibetan name ‘Yajima,’ growing in the highlands of China is a perennial herb belonging to the genus Chrysosplenium Saxifragaceae. As a traditional Chinese medicine, it has been used to treat digestive diseases for hundreds of years. The complete chloroplast genome of Chrysosplenium nudicaule is 152,775 bp in length and comprises two inverted repeats (IR, 25,962 bp), a large single-copy region (LSC, 83,533 bp), and a small single-copy region (SSC, 17,318 bp). It harbors 112 genes, including 78 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes. In addition, the rpl32 gene was deleted. The GC content of the whole chloroplast genome is 37.54%. This chloroplast genome resource will be useful for study on the evolution and genetic diversity of C. nudicaule in the future.
Chrysosplenium nudicaule Bunge is a small perennial herb belonging to the genus Chrysosplenium Saxifragaceae, and mainly distributed in the northwest and southwest mountain regions of China (Pan Citation1986). They grow in rock crevices ∼2500–4800 m above sea level. As a traditional Chinese medicine, it has been used to treat digestive diseases for hundreds of years in Tibet (Luo et al. Citation2016). A variety of chemical constituents were isolated from C. nudicaule, such as flavonoids and 78 volatile chemical constituents (Yang et al. Citation2004). The flavonoid compound, 6,7,3′-trimethoxy-3,5,4′-trihydroxy flavones isolated from C. nudicaule, can not only induce apoptosis in human leukemia K562 cells (Wang et al. Citation2005) but also inhibit growth and induce apoptosis of the human stomach cancer cell line SGC-7901 (Luo et al. Citation2016). Recent studies have also shown that the extract of this herb had a protective effect on liver injury in mice (Zhou et al. Citation2019). However, there are no reports on the molecular biology of C. nudicaule. In this study, we assembled and annotated its complete chloroplast genome from Illumina sequencing data. Then, we reconstructed the phylogenetic tree of this genus to reveal its genetic relationships. These data will provide some genetic resources for the subsequent molecular biology studies and lay a foundation for the development of its protection and medicinal value.
The materials of C. nudicaule in this study were collected from Qilian Mountain National Nature Reserve, Tianzhu County, Gansu Province of China (37°03′37″N, 102°46′05″E, 3275 m above sea level). The voucher specimens were deposited at the Herbarium of South-Central University for Nationalities (HSN). The specimen accession number is CY20210603001. The whole genomic DNA was extracted by the CTAB method (Doyle and Doyle Citation1987). The short-insertion library (350 bp) was constructed and then sequenced to obtain 2 × 150bp paired-end data by using the Illumina NovaSeq 6000 platform at Majorbio Company (Shanghai, China). A total of 5 Gb raw reads were generated and low-quality sequences were filtered out. First, the clean data were quality-controlled by using FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, de novo assembly of the whole chloroplast genome by GetOrganelle v1.7.3 (Jin et al. Citation2020). Finally, the final assembly result is obtained, we check the accuracy of assembly results, the slimmed assembly graph and selected target assembly graph can be visualized by Bandage v0.8.1 (Wick et al. Citation2015) to assess the completeness of the final graph. In CPGAVAS2 (Shi et al. Citation2019) and PGA (Qu et al. Citation2019), the C. nudicaule chloroplast genome is annotated by using Chrysosplenium sinicum (MK814606) as a reference genome. The different annotations of protein-coding sequences were confirmed using BLASTx in NCBI. The length of the complete chloroplast genome of C. nudicaule (MZ424445) was 152,775 bp with an average GC content of 37.54%. The complete chloroplast genome has a typical quadripartite structure, including a large single-copy (LSC) region of 83,533 bp, a small single-copy (SSC) region of 17,318 bp separated, and two inverted repeated regions of 25,962 bp. The chloroplast genome harbors 112 genes, including 78 protein-coding genes, 30 transfer RNA genes, and four ribosomal RNA genes. Notably, the rpl32 gene was absent in the complete chloroplast genome of C. nudicaule, which also lacked in some other species of Chrysosplenium L.
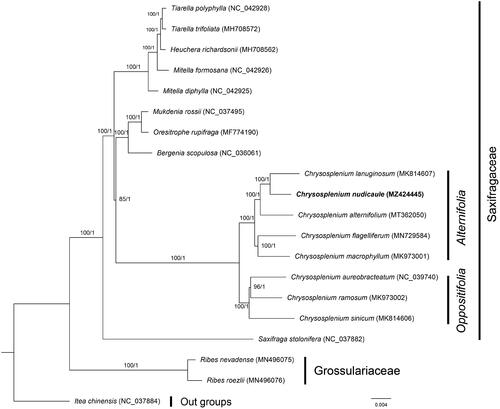
To further investigate the phylogenetic relationship of C. nudicaule within Saxifragaceae, the complete chloroplast genomes of nine species from Saxifragaceae, two species from Grossulariaceae and one species from Iteaceae were obtained from the GenBank database, with the Itea chinensis as the outgroups, the phylogenetic trees were built from the 78 protein-coding gene matrixes by maximum-likelihood (ML) and Bayesian inference (BI) (). The ML tree was generated using IQ-TREE (Nguyen et al. Citation2015) based on the best model of TVM + F + R2 with 1000 bootstrap replicates, and BI analysis was performed in MrBayes v3.2.6 (Ronquist et al. Citation2012). As shown in the phylogenetic tree (), we can see that the eight species of Chrysosplenium were clustered into a clade. In addition, they are divided into two major branches corresponding to the two subgenera (Alternifolia and Oppositifolia), and the result is consistent with those of Wu et al. (Citation2020). Chrysosplenium nudicaule belongs to the Subgen. Alternifolia, and has a close relationship with the Chrysosplenium lanuginosum. These studies will provide some references for the phylogeny, evolution, development, and utilization of Chrysosplenium in the future.
Acknowledgments
We acknowledge support from the teamwork project funded by National Forestry and Grassland Administration and Hubei Provincial Wildlife Conservation Station.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ424445. The associated ‘BioProject,’ ‘SRA,’ and ‘Bio-Sample’ numbers are PRJNA739074, SRR14861288, and SAMN19771098, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Luo Y, Yu H, Yang Y, Tian W, Dong K, Shan J, Ma X. 2016. A flavonoid compound from Chrysosplenium nudicaule inhibits growth and induces apoptosis of the human stomach cancer cell line SGC-7901. Pharm Biol. 54(7):1133–1139.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pan J-T. 1986. A study on the genus Chrysosplenium L. from China. J Univ Chinese Acad Sci. 24:81–97.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wang YP, Yang YS, Yang LX, Gao L, Zuo B. 2005. K562 apoptosis induced by flavone from Tibetan medicine Chrysosplenium nudicaule Bunge and its molecular mechanism. Pract J Cancer. (4):374–376.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Wu Z, Liao R, Yang T, Dong X, Lan D, Qin R, Liu H. 2020. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae). BMC Genomics. 21(1):621.
- Yang YS, He Z, Song AX, et al. 2004. Study on the chemical constituents of Tibetan medicine Chrysosplenium nudicaule Bunge. Nat Prod Res Dev. 4(4):294–296.
- Zhou YF, Zhong GY, Zhu JX, Wei RR, Wen L, Jiang W, Cao L, Ren G. 2019. Protective effects and action mechanism of extract from Tibetan medicine Yajima (Chrysosplenium nudicaule) on mice with intrahepatic cholestasis induced by ANIT. China J Chinese Mater Med. 44(5):1058–1063.