Abstract
The complete mitogenome of Fusarium oxysporum f. sp. albedinis (FOA), the causal agent of the destructive fusarium wilt in date palm, is sequenced and assembled. The circular mitogenome of isolate Foa44 is 51,601 bp in length and contains 26 transfer RNA (tRNA) genes, one ribosomal RNA (rRNA), and 28 protein-coding genes. A mitogenome-based phylogenetic analysis of Fusarium revealed that FOA is congruent with previous nuclear-gene phylogenetic results.
Introduction
Fusarium oxysporum f. sp. albedinis (Killian & Maire) Malençon is a fungal pathogen causing Bayoud disease, also known as Fusarium wilt, on date palm (Phoenix dactylifera). This disease represents a major threat to the date palm industry in North African countries. In fact, Bayoud disease has decimated more than 10 million trees in Morocco over the last century (Djerbi Citation1982; Sedra M Citation2003; Sedra MH Citation2003a, Citation2003b; Sedra Citation2007). As a soil-borne pathogen, FOA spores and mycelium colonize date palm roots, spread internally through the vascular system, and cause external symptoms characterized by both external hemiplegia character and dried palm leaves having the appearance of wet feathers and ultimately resulting in date palm death (Bouhssini and Faleiro Citation2018). The nuclear genome of F. oxysporum f. sp. albedinis has been recently sequenced and assembled (Khayi et al. Citation2020). To understand the mitogenomic background of F. oxysporum f. sp. albedinis, we present here, for the first time, the complete mitochondrial (MT) genome of FOA. The Fusarium oxysporum f. sp. albedinis strain Foa44, originally isolated in 1999, from an infected date palm in Tafilalt-Rissani, Morocco, and deposited in the Moroccan Coordinated Collections of Microorganisms (CCMM) ([email protected]) and the Belgian Coordinated Collections of Microorganisms (BCCM) under the accession numbers MUCL 41814 and Foa44, respectively.
Total genomic DNA of Foa44 strain was extracted from freeze-dried mycelium using the cetyltrimethylammonium bromide (CTAB) method (Möller et al. Citation1992). A paired-end library was prepared using a Nextera DNA Flex library kit from total genomic DNA (0.5 μg), following the manufacturer’s protocol. The library was sequenced (2 × 150 bp) on a NovaSeq 6000 platform (Illumina, San Diego, CA). Adapters and low-quality reads were removed using CLC Genomics Workbench V12.
De novo assembly was performed using MaSurca V3.4 with default parameters (Zimin et al. Citation2013). The obtained assembly was subject to BLASTN searches against the reference mitochondrial genome sequence for Fusarium oxysporum f. sp. lycopersici (GenBank accession number CM010346). The BLASTN output identified a single homologous contig of 67,535 bp in length. This contained an ∼14 kbp flanking inverted repeat region, as determined by dot plot analysis in CLC genomics Workbench. The mitogenome sequence was circularized by fragmenting the contig in half and then assembling the two fragments using CLC Genomics Workbench. The raw reads were remapped on the assembled sequence to correct any conflicts created during the assembly step. The final length of FOA mitogenome is 51,601 bp with 31% in G + C content. The mitogenome of Foa44 was deposited at GenBank under accession number MW493386. The annotation was performed using GeSeq (Tillich et al. Citation2017) and MFannot pipelines (https://github.com/BFL-lab/Mfannot). The resulting annotations were manually inspected and curated in comparison to published F. oxysporum mitogenomes. In total, 28 protein-coding genes, one ribosomal RNA (rRNA), and 26 transfer RNA (tRNA) genes were predicted. Among 28 predicted genes, 14 genes are commonly found in MT genomes including those implicated in ATP production (atp6, atp8, and atp9), oxidative phosphorylation (nad1–6 and nad4L), apocytochrome b (cob), and cytochrome C oxidase subunits (cox1–3). The 14 remaining genes are of unknown functions except for four genes (orf529, orf304, orf292, and orf348) that are coding for Homing Endonucleases (GIY-YIG and LAGLIDADG) that are commonly found in fungal MT genomes (Megarioti and Kouvelis Citation2020).
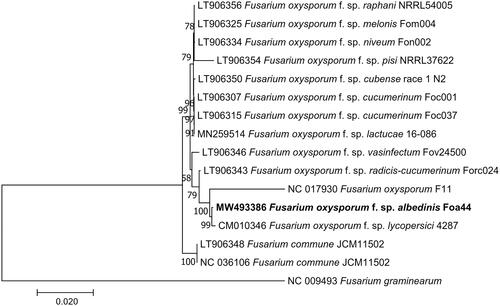
To highlight the phylogenetic position of F. oxysporum f. sp. albedinis Foa44 within Fusarium oxysporum complex, a whole MT genome alignment of 16 Fusarium mitogenome sequences retrieved from Genbank, was performed with MAFTT software (Katoh and Standley Citation2013). The Maximum-likelihood phylogenetic tree was constructed using MEGA7, under a Kimura 2-parameter model (Kumar et al. Citation2016). The resulting phylogenetic tree shows that our mitogenome is clearly clustered with two other strains F. oxysporum f. sp. lycopersici 4287 (CM010346) and F. oxysporum F11 (NC017930) (). Variant calling showed that there are 369 and 500 SNP/Indels differentiating the FOA mitogenome from these related mitogenomes, respectively.
Figure 1. Phylogenetic tree of Fusarium constructed using Maximum Likelihood method based on alignment of 16 whole mitochondrial genomes of F. oxysporum f. sp. albedinis (MW493386, this study), F. oxysporum f. sp. lactucae (MN259514), F. oxysporum (NC_017930), F. oxysporum f. sp. melonis (LT906325), F. oxysporum f. sp. pisi (LT906354), F. oxysporum f. sp. vasinfectum (LT906346), F. oxysporum f. sp. niveum (LT906334), F. oxysporum f. sp. cucumerinum (LT906307), F. oxysporum f. sp. radicis-lycopersici (CM010346), F. commune (LT906348 and NC_036106), F. oxysporum f. sp. raphani (LT906356), F. oxysporum f. sp. cubense (LT906350), F. oxysporum f. sp. cucumerinum (LT906315), F. oxysporum f. sp. radices-cucumerinum (LT906343), and F. graminearum (NC_009493) as an outgroup). The tree with the highest log likelihood (−64,062.86) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with a superior log-likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 27,505 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. Citation2016).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW493386. The associated **BioProject**, **BioSample**, and **SRA** numbers are PRJNA658960, SAMN15893572, and SRP313423, respectively.
References
- Bouhssini ME, Faleiro JR. 2018. Date palm pests and diseases. International Center for Agricultural Research in the Dry Areas (ICARDA). Lebanon.
- Djerbi M. 1982. Bayoud disease in North Africa [date palms, fungus disease]: history, distribution, diagnosis and control. Date Palm J. 1(2):153–197.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khayi S, Khoulassa S, Gaboun F, Abdelwahd R, Diria G, Labhilili M, Iraqi D, El Guilli M, Fokar M, Mentag R. 2020. Draft genome sequence of Fusarium oxysporum f. sp. albedinis strain Foa 133, the causal agent of Bayoud disease on date palm. Microbiol Resour Announc. 9:e00462-20. doi:10.1128/MRA.00462-20.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Megarioti AH, Kouvelis VN. 2020. The coevolution of fungal mitochondrial introns and their homing endonucleases (GIY-YIG and LAGLIDADG). Genome Biol Evol. 12(8):1337–1354.
- Möller EM, Bahnweg G, Sandermann H, Geiger HH. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20(22):6115–6116.
- Sedra M. 2003. Date palm cultivation, characterization and classification of main Mauritanian varieties. Al-Khartoum: AOAD.
- Sedra M. 2007. Bayoud disease of date palm in North Africa: recent distribution and remarks about its characterization, diagnosis and origin. Proceedings Fourth Symposium Date Palm. Hofuf: King Faisal University.
- Sedra MH. 2003a. Le bayoud du palmier dattier en Afrique du Nord. Tunis (Tunisia): Bureau Sous-Regional pour l’Afrique du Nord.
- Sedra MH. 2003b. Le palmier dattier base de la mise en valeur des oasis au Maroc. Maroc: INRA.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics. 29(21):2669–2677.
