Abstract
Vespa magnifica (Smith) is an aggressive social wasp species of Vespidae family. This species is of medical importance for its dangerous sting, traditional medicinal use and valuable venom components. Here, a complete mitogenome of V. magnifica was presented. It was 16,730 bp in length with nucleotide composition of AT: 79.4% and CG: 20.6%. In total, 13 protein-coding genes (PCGs), 22 transfer RNA, and two ribosomal RNA genes were annotated in this mitogenome. Phylogenetic analysis was performed using V. magnifica with 20 other species of Vespidae. The result indicated those species of genus Vespa fell into a paraphyletic group. Moreover, the Vespa species with large body size were clustered into a clade. This mitogenome resource can contribute to further phylogenetic and taxonomic study on genus Vespa.
Keywords:
Stings of wasps are common outdoor risks to people who disturb those aggressive insects deliberately or accidentally. Clinical syndromes following the sting are often characterized by itching, swelling, and acute pain. Allergic reactions with serious clinical outcomes are also commonly reported from emergency department globally. Vespa magnifica (Vespidae) is an aggressive wasp species with large body size. Cases of serious syndrome after sting of this species were reported from Asia countries (Vikrant et al. Citation2005; George et al. Citation2008; Vikrant and Parashar Citation2017). Meanwhile, the raw venom of V. magnifica is a traditional medicine for rheumatoid arthritis, which was employed by Jingpo ethnic group of Yunnan Province, China (Zhou et al. Citation2019). Many researches had revealed valuable components from the venom of V. magnifica, such as bioactive peptide, anticoagulant serine protease, kininogen, etc (Xu et al. Citation2006; Han et al. Citation2008; An et al. Citation2012). In Yunnan Province of China, V. magnifica together with other species Vespa mandarinia and Vespa ducalis, were all named as ‘da tu feng,’ which particularly emphasizes their large body size, dangerous sting, as well as similar morphological characters. Considering the potentially medical importance of V. magnifica, a complete mitochondrial genome sequence of V. magnifica (GenBank: MT137097.2) was provided here.
The female V. magnifica samples were captured by Zichao Liu in Xinzhai Village, Lvchun County, Yunnan Province, China (102°50′N, 22°80′E). The wasps were attracted by baits of chicken meat and captured by insect nets. Then they were deposited into pure ethyl alcohol for storage. The species identification was performed following the description of its morphological characters (Rao et al. Citation2014). The voucher specimen of V. magnifica used in this study was assigned with a unique series code (MG20201101-3) and deposited into the bio-sample herbarium of Institute of Emergency and Critical Care Medicine of Changsha. The thorax part of wasp was detached using cleaned fine forceps and muscle of voucher sample was transferred to a DNAase-free tube. Then raw DNA material was extracted using TIANamp Genomic DNA Kit (DP304) (TIANGEN BIOTECH, Beijing, China). The DNA material was processed and qualified following the instruction of Illumina sequencing platform. The PE150 libraries were prepared using TruSeq Nano DNA LT Sample Preparation Kit (Illumina, San Diego, CA, USA).
Sequencing was performed on the Illumina HiSeq X Ten platform (Illumina Inc., San Diego, CA, USA) using the prepared DNA libraries. After sequence filtering by SOAPnuke (version: 1.3.0) (Chen et al. Citation2018), 2.5 Gb clean reads were finally assembled into a complete circular mitochondrial genome of 16,730 bp in length using MITObim (v1.8) (Hahn et al. Citation2013). The sequence was deposited into GenBank and given a unique accession number MT137097.2. The nucleotide composition was calculated using MEGA 7 software, which showed AT: 79.4% and CG: 20.6% (T:40.7%, A:38.7%, C:14.7%, G:5.9%) (Kumar et al. Citation2016). The protein-coding gene (PCGs) annotation and the prediction of tRNAs secondary structure (except for tRNA-Asn and tRNA-Ser) were performed by applying Mitochondrial Genome annotation 2 (MITOS2) webserver (Bernt et al. Citation2013). In total, 13 PCGs, 22 tRNA, and two rRNA genes were annotated in this mitogenome. The open reading frames (ORF) features of all the 13 PCGs were recognized using online version of ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/).
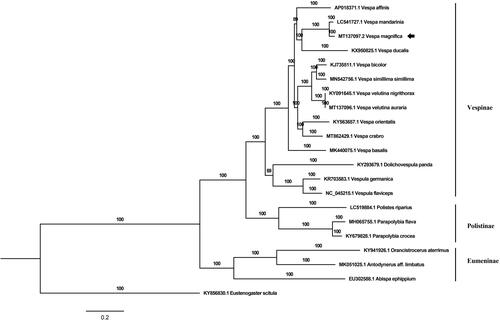
For phylogenetic analysis, a Bayesian tree was constructed based on PCGs and two rRNA genes of V. magnifica and 20 other species of Vespidae. The MrBayes (v3.2.4) was used for this analysis. The chain length was set as 5,000,000 generations and sampled every 1000 generations during the calculation (Ronquist et al. Citation2012). Eustenogaster scitula was set as outgroup (). The result of phylogenetic analysis was largely consistent with previous reports (Kim et al. Citation2017; Liu et al. Citation2020). All the species of genus Vespa fell into a large group which was paraphyletic. In addition, the species with large body sizes, including V. magnifica, V. mandarinia, and V. ducalis, were clustered into a clade. The present mitogenome of V. magnifica could contribute to the phylogenetic studies of the genus Vespa and the identification of large wasp species identification.
Acknowledgments
The authors thank FM Meng ([email protected]) and ZC Liu ([email protected]) for sample collection and permission to use the mitochondrial resource in the present paper.
Disclosure statement
All authors announce no conflicts of interest in the present paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MT137097.2. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA703057, SRS8300133 and SAMN18011289 respectively.
Additional information
Funding
References
- An S, Chen L, Wei J-F, Yang X, Ma D, Xu X, Xu X, He S, Lu J, Lai R, et al. 2012. Purification and characterization of two new allergens from the venom of Vespa magnifica. PLoS One. 7(2):e31920.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- George P, Pawar B, Calton N, Mathew P. 2008. Wasp sting: an unusual fatal outcome. Saudi J Kidney Dis Transpl. 19(6):969–972.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Han J, You D, Xu X, Han W, Lu Y, Lai R, Meng Q. 2008. An anticoagulant serine protease from the wasp venom of Vespa magnifica. Toxicon. 51(5):914–922.
- Kim JS, Jeong JS, Kim I. 2017. Complete mitochondrial genome of the yellow-legged Asian hornet, Vespa velutina nigrithorax (Hymenoptera: Vespidae). Mitochondrial DNA B Resour. 2(1):82–84.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu Z, Tang H, Tong L, Liu X, Meng F. 2020. The complete mitochondrial genome of a forensic potential wasp, Vespa auraria (Smith). Mitochondrial DNA B Resour. 5(3):3455–3456.
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159(7):1665–1680.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Vikrant S, Pandey D, Machhan P, Gupta D, Kaushal SS, Grover N. 2005. Wasp envenomation-induced acute renal failure: a report of three cases. Nephrology. 10(6):548–552.
- Vikrant S, Parashar A. 2017. Two cases of acute kidney injury due to multiple wasp stings. Wilderness Environ Med. 28(3):249–252.
- Xu X, Li J, Lu Q, Yang H, Zhang Y, Lai R. 2006. Two families of antimicrobial peptides from wasp (Vespa magnifica) venom. Toxicon. 47(2):249–253.
- Zhou S-T, Luan K, Ni L-L, Wang Y, Yuan S-M, Che Y-H, Yang Z-Z, Zhang C-G, Yang Z-B. 2019. A strategy for quality control of Vespa magnifica (Smith) venom based on HPLC fingerprint analysis and multi-component separation combined with quantitative analysis. Molecules. 24(16):2920.