Abstract
Sparganium stoloniferum subsp. choui is an aquatic perennial herb distributed in Northeast China. It was published as a new species in 1992 and recognized as a subspecies of S. stoloniferum in Flora of China in 2010. The complete chloroplast genome of S. stoloniferum subsp. choui was sequenced and assembled. The genome size was 161,865 bp in length with 36.8% GC content. Its quadripartite structure consisted of the large single copy (LSC, 88,953 bp) and small single copy (SSC, 19,098 bp) regions, separated by a pair of inverted repeats (IRs) of 26,907 bp. The genome contained 114 distinct genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis including four Sparganium species showed that two subspecies of S. stoloniferum were not monophyletic, supporting the resurrection of S. choui as a species.
Sparganium L. is an aquatic perennial genus including approximately 14–19 species and occurs mainly in temperate and cool regions (Cook and Nicholls Citation1986, Citation1987; Kaul Citation2000; Sun and Simpson Citation2010). Sparganium stoloniferum (Buch.-Ham. ex Graebn.) Buch.-Ham. ex Juz. is a widespread species in Asia and of ecological and economic importance. The dried tuberous rhizome of S. stoloniferum is a widely used gynecological drug in traditional Chinese medicine (Jia et al. Citation2021). Two subspecies of S. stoloniferum, subspecies stoloniferum and Sparganium stoloniferum subsp. choui (D.Yu) K.Sun, were recorded in Flora of China (Sun and Simpson Citation2010). The latter was recognized from a new species S. choui published by Yu (Citation1992), which differs from S. stoloniferum among panicle length, male head number, anther length, and fruit size (Yu Citation1992; Sun and Simpson Citation2010). The complete chloroplast (cp) genome of S. stoloniferum has been reported (Su et al. Citation2019). Here, we reported complete cp genome of S. stoloniferum subsp. choui and constructed phylogenetic tree to test the monophyly of two subspecies of S. stoloniferum.
The sample of S. stoloniferum subsp. choui was collected from Arong Banner, Inner Mongolia, China (48.13°N, 123.46°E). A specimen was deposited at the herbarium of Wuhan University (www.whu.edu.cn, Xinwei Xu, [email protected]) under the voucher number Xu568. Total genomic DNA was extracted by using the DNA Secure Plant Kit (Tiangen Biotech, Beijing, China). Library preparation and genome sequencing were conducted on the Illumina Hiseq 2500 platform (Benagen Inc., Wuhan, China). SPAdes v.3.9.0 (Nurk et al. Citation2017) was used to assemble the obtained paired-end reads. With the reference cp genome of Typha latifolia L. (Guisinger et al. Citation2010; NC_013823.1), the assembled sequence was annotated in GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017). The obtained complete cp genome of S. stoloniferum subsp. choui was then submitted to GenBank (accession no. MW829765).
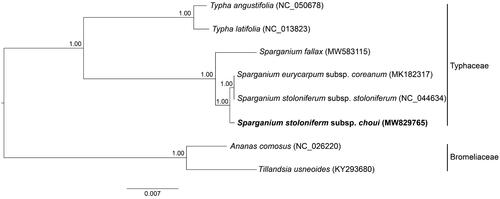
The complete cp genome of S. stoloniferum subsp. choui is 161,865 bp in length, including a large single copy (LSC) region of 88,953 bp, a small single copy (SSC) region of 19,098 bp, and two inverted repeat (IR) regions of 26,907 bp. The GC contents of LSC, SSC, IR and whole genome are 34.8%, 30.4%, 42.4%, and 36.8%, respectively. There are 114 distinct genes annotated, including 80 protein coding genes, 30 tRNA genes, and four rRNA genes. A total of 18 genes contain introns, and three of them have two introns (ycf3, clpP, and rps12). Bayesian inference of phylogeny was conducted based on eight cp genomes of Typhaceae and Bromeliaceae species using MrBayes v.3.2.7a (Ronquist et al. Citation2012). The phylogenetic tree showed that two subspecies of S. stoloniferum was not monophyletic, and subspecies stoloniferum was most closely related to S. eurycarpum rather than subspecies choui (), supporting the resurrection of S. choui as a species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW829765. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA718700, SRR14126901, and SAMN1855935, respectively.
Additional information
Funding
References
- Cook CDK, Nicholls MS. 1986. A monographic study of the genus Sparganium (Sparganiaceae). Part 1. Subgenus Xanthosparganium Holmberg. Bot Helv. 96:213–267.
- Cook CDK, Nicholls MS. 1987. A monographic study of the genus Sparganium (Sparganiaceae). Part 2. Subgenus Sparganium. Bot Helv. 97:1–44.
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70(2):149–166.
- Jia J, Li X, Ren X, Liu X, Wang Y, Dong Y, Wang X, Sun S, Xu X, Li X, et al. 2021. Sparganii Rhizoma: a review of traditional clinical application, processing, phytochemistry, pharmacology, and toxicity. J Ethnopharmacol. 268:113571.
- Kaul RB. 2000. Sparganiaceae. In: Flora of North America Editorial Committee, editor. Flora of North America North of Mexico. Vol. 22: Magnoliophyta: Alismatidae, Arecidae, Commelinidae (in Part) and Zingiberidae. New York and Oxford: Oxford University Press; p. 270–277.
- Nurk S, Meleshko D, Korobeyniko A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Su T, Yang JX, Lin YG, Kang N, Hu GX. 2019. Characterization of the complete chloroplast genome of Sparganium stoloniferum (Poales: Typhaceae) and phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1402–1403.
- Sun K, Simpson DA. 2010. Typhaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 23. Acoraceae through Cyperaceae. Beijing (China)/St. Louis (MI): Science Press/Missouri Botanical Garden Press; p. 158–163.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu D. 1992. New plants of Sparganium in Northeastern China. Bull Bot Res. 12(3):255–261.