Abstract
The peacock butterfly Aglais io (Linnaeus, 1758) (Nymphalidae: Nymphalinae: Nymphalini) is a colorful and charismatic flagship butterfly species whose range spans from the British Isles and Europe through temperate Asia and the Far East. In Europe, it has been used as a model species for studying the effects of GMO maize pollen on caterpillar growth and survivorship. The Japanese subspecies, Aglais io geisha (Stichel Citation1907), is not as well studied as its European counterpart. Genome skimming by Illumina sequencing allowed the assembly of a complete circular mitochondrial genome (mitogenome) of 15,252 bp from A. io geisha consisting of 80.6% AT nucleotides, 13 protein-coding genes, 22 tRNAs, two rRNAs, and a control region in the gene order typical of butterfly species. Aglais io geisha COX1 gene features an atypical start codon (CGA) while COX1, COX2, CYTB, ND1, ND3, ND4, and ND5 display incomplete stop codons finished by the addition of 3’ A residues to the mRNA. Bayesian phylogenetic reconstruction places A. io geisha within a clade with European A. io mitogenomes in the tribe Nymphalini, which is consistent with previous phylogenetic hypotheses.
The peacock butterfly Aglais io (Linnaeus, 1758) (Nymphalidae: Nymphalinae: Nymphalini) is an indicator species for studying the effects of GMO maize pollen on non-targeted insects in Europe (Arpaia et al. Citation2018; Leclerc et al. Citation2018). The natural range of A. io includes the British Isles, Europe, temperate Asia, and the Far East, but has recently expanded its range into North America (Nazari et al. Citation2018).
Aglais io is a colorful bivoltine species producing two broods of offspring, with one that flies in summer and one that over-winters as adults (Arpaia et al. Citation2018; Leclerc et al. Citation2018). The adults feed on nectar-bearing plants, while the caterpillars feed on members of the nettle family Urticaceae which is the route by which GMO maize pollen is ingested (Leclerc et al. Citation2018). Adult females lay eggs in large pyramidal clusters on hops plants (Humulus lupulus) to protect the innermost layer of eggs from parasitism by flies (Tachnidae) and wasps (Ichneumonidae) (Hondo et al. Citation1995; Audusseau et al. Citation2021). Adults have been observed to fake death upon wings being pinched together, with antennae becoming immobile and legs stiffening against the body (Loxdale Citation2017). Adults also have sound-producing eye-spots on their wings to deter predators including bats and birds (Møhl and Miller Citation1976; Vallin et al. Citation2005; Loxdale Citation2017). Less is known about the focus of the current study, the Japanese peacock butterfly, subspecies A. io geisha (Stichel Citation1907), than its European counterpart A. io.
Here we report the complete mitochondrial genome (mitogenome) sequence of A. io geisha from specimen Ai2015.2, collected in Saitama, Japan (GPS 35.90807 N, 139.65657E) in July 2015 that has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (http://www.wallisroughley.ca/, Jason Gibbs, [email protected]) voucher WRME0507739.
DNA was prepared (McCullagh and Marcus Citation2015) and later sequenced by Illumina NovaSeq6000 (San Diego, CA) (Marcus Citation2018). The mitogenome of A. io geisha (Genbank MZ322948) was assembled and annotated using Geneious Prime 2021.1 from an SRA library of 23,191,042 paired 150 bp reads (Genbank SRA PRJNA733565) using Aglais io and Araschnia levana reference mitogenomes (Lepidoptera: Nymphalidae, KM592970; Lepidoptera: Nymphalidae, MT712075) (Timmermans et al. Citation2016; Alexiuk et al. Citation2020a). The A. io geisha nuclear rRNA repeat (Genbank MZ322949) was also assembled and annotated using an A. levana (MT750296) reference sequence. The rRNA repeat sequence is increasingly recognized as being very useful for phylogenetic comparisons based on nuclear markers (Dodsworth Citation2015; Coissac et al. Citation2016; Marcus Citation2018; Krehenwinkel et al. Citation2019), so we have chosen to release it here.
The A. io geisha circular 15,252 bp mitogenome assembly was composed of 2700 paired reads with nucleotide composition: 40.1% A, 11.9% C, 7.5% G, and 40.5% T. The gene composition and order in A. io geisha is typical of the arrangement found in most butterfly mitogenomes (Park et al. Citation2016). The A. io geisha protein-coding gene start codons include ATG (ATP6, COX2, COX3, CYTB, ND1, ND4), ATT (ND2, ND3, ND5), ATC, (ND6), CGA, an atypical COX1 start codon that is also found in the COX1 gene of many other insects (Liao et al. Citation2010). Additionally, ATP8 and ND4L have ATA start codons that are infrequently used in insect mitochondria but are frequently used in other animal groups (Okimoto et al. Citation1990; Han et al. Citation2016; Alexiuk et al. Citation2020b). The mitogenome contains five protein-coding genes (COX1, COX2, CYTB, ND3, ND5) with single-nucleotide (T––) stop codons, and two protein-coding genes (ND1, ND4) with two-nucleotide (TA–) stop codons completed by post-transcriptional addition of 3′ A residues. All structures of the tRNAs were verified using ARWEN v.1.2 (Laslett and Canback Citation2008) and have typical cloverleaf secondary structures with the exception for trnS (AGN) where the dihydrouridine arm is replaced by a loop, whereas the control region and mitochondrial rRNAs are typical for Lepidoptera (McCullagh and Marcus Citation2015).
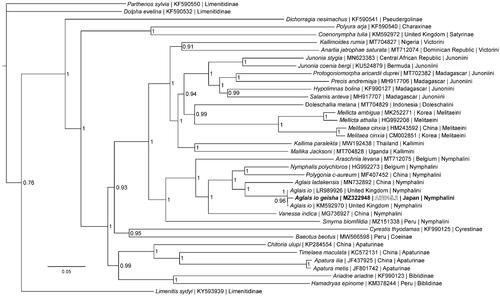
Phylogenetic reconstruction () was completed using the complete mitogenome of A. io geisha, 34 mitogenomes from the subfamily Nymphalinae and three outgroup species from subfamily Limenitidinae (Alexiuk et al. Citation2020b; Hamilton et al. Citation2020; Lalonde and Marcus Citation2020; Payment et al. Citation2020; Lalonde Citation2021; Alexiuk et al. Citation2021). Mitogenome sequences were aligned in CLUSTALX 2.1 (Thompson et al. Citation1997; Larkin et al. Citation2007) and analyzed using Bayesian Inference with the GTR + I + G model (model selected using jModeltest 2.1.1 (Darriba et al. Citation2012)) in Mr. Bayes version 3.2.7 (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012). As expected based on previous phylogenetic hypotheses (Nylin et al. Citation2001; Wahlberg and Nylin Citation2003; Wahlberg et al. Citation2009), phylogenetic analysis places A. io geisha in a clade with mitogenomes from European A. io specimens within tribe Nymphalini.
Figure 1. The Bayesian phylogeny (GTR + I + G model, average Potential Scale Reduction Factor (PSRF) = 1, average deviation of split frequencies = 0.000628) of the Aglais io geisha mitogenome, 37 additional mitogenomes from within family Nymphalidae, including outgroup species Limenitis sydyi, Parthenos sylvia, and Dophla evelina (Limenitinae) (Alexiuk et al. Citation2020b; Hamilton et al. Citation2020; Lalonde and Marcus Citation2020; Payment et al. Citation2020; Lalonde Citation2021), produced by 10 million MCMC generations in MrBayes, with sampling every 100 generations, and after discarding the first 250,000 generations as burn-in. The Bayesian posterior probability values determined by MrBayes are provided at each node.
Acknowledgments
Thanks to Genome Quebec for assistance with library preparation and sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession nos. MZ322948 and MZ322949. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA733565, SRX11064013, and SAMN19415664 respectively.
Additional information
Funding
References
- Alexiuk MR, Lalonde MML, Marcus JM. 2021. Phylogenetic analysis of the complete mitochondrial genome of the Blomfild’s Beauty butterfly Smyrna blomfildia (Fabricius 1781) (Insecta: Lepidoptera: Nymphalidae: Nymphalini). Mitochondrial DNA B Resour.
- Alexiuk MR, Marcus JM, Lalonde MML. 2020a. The complete mitochondrial genome and phylogenetic analysis of the European map butterfly Araschnia levana (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5(3):3246–3266.
- Alexiuk MR, Marcus JM, Lalonde MML. 2020b. The complete mitochondrial genome of the Jackson's leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5(3):3298–3318.
- Arpaia S, Baldacchino F, Bosi S, Burgio G, Errico S, Magarelli RA, Masetti A, Santorsola S. 2018. Evaluation of the potential exposure of butterflies to genetically modified maize pollen in protected areas in Italy. Insect Sci. 25(4):549–561.
- Audusseau H, Ryrholm N, Stefanescu C, Tharel S, Jansson C, Champeaux L, Shaw MR, Raper C, Lewis OT, Janz N, et al. 2021. Rewiring of interactions in a changing environment: nettle-feeding butterflies and their parasitoids. Oikos. 130(4):624–636.
- Coissac E, Hollingsworth PM, Lavergne S, Taberlet P. 2016. From barcodes to genomes: extending the concept of DNA barcoding. Mol Ecol. 25(7):1423–1428.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dodsworth S. 2015. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 20(9):525–527.
- Hamilton RV, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5(3):3306–3308.
- Han Z, Wang G, Xue T, Chen Y, Li J. 2016. The F-type complete mitochondrial genome of Chinese freshwater mussels Cuneopsis pisciculus. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3376–3377.
- Hondo M, Onodera T, Morimoto N. 1995. Parasitoid attach on a pyramid-shaped egg mass of the peacock butterfly, Inachis io geisha (Lepidoptera: Nymphalidae). Appl Entomol Zool. 30(2):271–276.
- Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG, et al. 2019. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. GigaScience. 8(5):giz006.
- Lalonde MML. 2021. Phylogenetic analysis of the complete mitochondrial genome of the graphic beauty butterfly Baeotus beotus (Doubleday 1849) (Lepidoptera: Nymphalidae: Nymphalinae: Coeini). Mitochondrial DNA B Resour. 6(4):1516–1518.
- Lalonde MML, Marcus JM. 2020. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniomorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5(3):3243–3263.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Leclerc M, Walker E, Messéan A, Soubeyrand S. 2018. Spatial exposure-hazard and landscape models for assessing the impact of GM crops on non-target organisms. Sci Total Environ. 624:470–479.
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186.
- Loxdale HD. 2017. Butterflies playing 'possum': an adaptive behaviour related to winter survival. Antenna. 41:11–16.
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18(4):749–755.
- Møhl B, Miller LA. 1976. Ultrasonic clicks produced by the peacock butterfly: a possible bat-repellent mechanism. J Exp Biol. 64(3):639–644.
- Nazari V, Handfield L, Handfield D. 2018. The European peacock butterfly, Aglais io (Linnaeus 1758) in North America (Lepidoptera: Nymphalidae). News Lep Soc. 60:128–129.
- Nylin S, Nyblom K, Ronquist F, Janz N, Belicek J, Källersjö M. 2001. Phylogeny of Polygonia, Nymphalis and related butterflies (Lepidoptera: Nymphalidae): a total-evidence analysis. Zool J Linn Soc. 132(4):441–468.
- Okimoto R, Macfarlane JL, Wolstenholme DR. 1990. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 18(20):6113–6118.
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826.
- Payment JE, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resources. 5:3415–3417.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stichel H. 1907. Brassoliden-Studien. Mitt Mus Natkd Berl, Dtsch Entomol. 1907(10):160–179.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Timmermans MJTN, Viberg C, Martin G, Hopkins K, Vogler AP. 2016. Rapid assembly of taxonomically validated mitochondrial genomes from historical insect collections. Biol J Linn Soc. 117(1):83–95.
- Vallin A, Jakobsson S, Lind J, Wiklund C. 2005. Prey survival by predator intimidation: an experimental study of peacock butterfly defence against blue tits. Proc Biol Sci. 272(1569):1203–1207.
- Wahlberg N, Nylin S. 2003. Morphology versus molecules: resolution of the positions of Nymphalis, Polygonia and related genera (Lepidoptera: Nymphalidae). Cladistics. 19(3):213–223.
- Wahlberg N, Weingartner E, Warren A, Nylin S. 2009. Timing major conflict between mitochondrial and nuclear genes in species relationships of Polygonia butterflies (Nymphalidae: Nymphalini). BMC Evol Biol. 9:92.
