Abstract
In this study, we presented the first complete mitochondrial genome of the genus Simocephalus determined by next-generation sequencing. The mitogenome of S. sibiricus is 15,818 bp in length, including 13 protein-coding genes (PCGs), two ribosomal RNAs, 22 tRNAs, and one putative control region, and has the same gene order with Daphnia. ATG and ATT were used as start codons in 11 PCGs, TTG was used in COX1 and GTG in ATP8. Six PCGs used an incomplete stop codon. Phylogenetic analysis based on 13 PCGs showed that, on genus level, Simocephalus was closely related to Daphnia.
The genus Simocephalus is a large group in Cladocera, commonly occurring in littoral zones of ponds and lakes. It was widely distributed but less studied (Forró et al. Citation2008; Xiang et al. Citation2015). The genus Simocephalus was reviewed by Orlova-Bienkowskaja (Citation2001) and grouped into five subgenera: Simocephalus s. str., Acutirostratus, Aquipiculus, Coroncephalus and Echinocaudus. These subgenera were widely accepted by taxonomists, but many species in the genus Simocephalus were not (Kotov et al. Citation2013), e.g., S. himalayensis Chiang and Chen, 1974 and S. himalayensis microdus Chen, Shi and Shi, 1992. There were 14 species of Simocephalus recorded in China, but three of them were in the state of incertae sedis and two of them were considered as synonyms (Xiang et al. Citation2015). The first record of S. (E.) sibiricus Sars, 1898 in China was reported by Shi and Shi (Citation1994) and distributed only in Heilongjiang Province (Shi and Shi Citation1996), which was established as a synonym of S. exspinosus later by Orlova-Bienkowskaja (Citation2001). However, the validity of S. sibiricus was confirmed by genetic sequences (using COXI and 18S sequences) and some morphological characters (short rhomboid ocellus, with 16–22 spines on the basal postabdomen claw, and the dorsal and ventral posterior valve margin with thick and strong denticles) (Huang et al. Citation2014). At present, there are only two mitogenomes of Simocephalus that were published on the NCBI database, i.e., one annotated partial mitogenome of S. vetulus (MT862434) and one non-annotated mitogenome of S. serrulatus (LS991523). In this study, we sequenced and annotated one complete mitogenome of S. sibiricus from southern China. It is also the first complete mitogenome for the genus Simocephalus, which will contribute to further study of the genus Simocephalus and the phylogeny of the Daphnidae.
The specimens of Simocephalus were collected in March 2021 from a pond in South China Botanical Garden (113.37°E, 23.19°N), Guangzhou City, southern China. The individuals of Simocephalus were identified as S. sibiricus with morphological characters of short rhomboid ocellus, around 20 spines on the basal postabdomen, thick and strong denticles on the dorsal ventral posterior valve margin according to Orlova-Bienkowskaja (Citation2001) and Huang et al. (Citation2014). The specimens of S. sibiricus were deposited at the Animal Specimens Museum of Jinan University (https://hydrobio.jnu.edu.cn/2021/0316/c29093a602403/page.htm, contact person: Ningning Liu; email: [email protected]) under the voucher number ZOOPA07005. Around 150 specimens were picked out to extract the total DNA by using the TIANamp Marine Animals DNA Kit. The DNA was also stored at −80 °C at the Animal Specimens Museum of Jinan University. Genomic library was constructed by a high-throughput sequencing technology on Illumina's Novaseq family of platforms. We used the library and splicing software Novoplasty (https://github.com/ndierckx/NOVOPlasty) to conduct the de novo assembly of mitogenome with a COI sequence (KF960086) of S. sibiricus as seed. The annotation was based on the comparison annotation of NCBI mitochondrial database, with mitogenome of S. vetulus (GenBank Accession Number: MT862434) as seed.
The mitogenome of S. sibiricus (NO. MW848816) was 15,818 bp in length, containing 13 PCGs, two ribosomal RNA genes (rrnL and rrnS), 22 transfer RNA genes (tRNAs), and one putative control region. The gene order of S. sibiricus was consistent with that of daphnids. The nucleotide composition of the mitogenome has a significantly biased A + T content of 70.13% (A, G, C, and T was 33.93%, 14.30%, 15.57%, and 36.20%, respectively). The control region waslocated between tRNA-Ile and tRNA-Gln with a length of 1230 bp, and the A + T content was 73.17%. Four PCGs (ND1, ND4, ND4L, ND5), two rRNAs (rrnL, rrnS) and nine tRNAs (tRNA-Ile, tRNA-Gln, tRNA-Cys, tRNA-Tyr, tRNA-Phe, tRNA-His, tRNA-Pro, tRNA-Leu, and tRNA-Val) were transcribed from the L-strand, while the remaining 22 genes were encoded on the H-strand. ATG was used as the start codon in the most genes, i.e., ATP6, COX2, COX3, CYTB, ND2, ND4, and ND5. ATT was used as the start codon in ND1, ND3, ND4L, and ND6, TTG in COX1, and GTG in ATP8. Among 13 PCGs, seven ended up with typical stop codon of TAA or TAG, and six used an incomplete stop codon with single ‘T––’ (COX1, COX2, ND3, ND4, ND5 and CYTB).
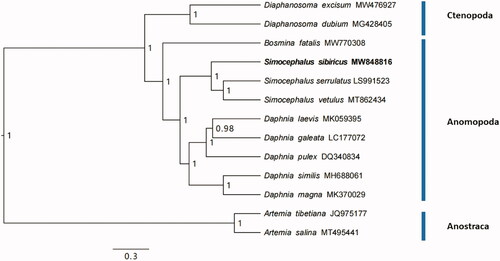
The phylogenetic tree was obtained using Bayesian Inference (BI) analysis based on entire protein coding genes of eleven Cladocera including the two Simocephalus species mentioned above (MT862434 and LS991523) and two outgroups (Artemia tibetiana and A. salina) in software BEAST v1.8.4, with GTR substitution model, MCMC chains of 100,000,000, and burn in 10%. The mitogenome of the S. sibiricus had similar A + T component with the other two Simocephalus species (69.2% for S. vetulus and 68.4% for S. serrulatus). The phylogenetic tree showed that the three Simocephalus species were in a cluster. On genus level, Simocephalus were closely related to Daphnia for the Daphniidae but was fully separated from Bosminidae (). At a deeper level, the genus Simocephalus (order Anomopoda) was phylogenetically closer to the Diaphanosoma (order Ctenopoda) but greatly divergent from Artemia (order Anostraca). The phylogenetic relationships among the three orders (Anostraca, Ctenopoda, Anomopoda) and within Anomopoda in our study were identical to that of Schwentner et al. (Citation2018) based on transcriptome data.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov) under the accession no. MW848816. The associated BioProject, SRA, and BioSample numbers are PRJNA719360, SRR1413763, and SAMN18603966 respectively.
Additional information
Funding
References
- Forró L, Korovchinsky NM, Kotov AA, Petrusek A. 2008. Global diversity of cladocerans (Cladocera; Crustacea) in freshwater. Hydrobiologia. 595(1):177–184.
- Huang XN, Shi XL, Kotov AA, Gu FK. 2014. Confirmation through genetic analysis of the existence of many local phyloclades of the genus Simocephalus (Crustacea, Cladocera) in China. PLoS One. 9(11):e112808.
- Kotov AA, Korovchinsky NM, Petrusek A. 2013. World checklist of freshwater Cladocera species. Available from: http://fada.biodiversity.be/group/show/17
- Orlova-Bienkowskaja MY. 2001. Cladocera, Anomopoda: Daphniidae, genus Simocephalus. Leiden: Backhuys; p. 130.
- Schwentner M, Richter S, Rogers DC, Giribet G. 2018. Tetraconatan phylogeny with special focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics. Proc R Soc B. 285:20181524.
- Shi XL, Shi XB. 1994. On two new species and two new records of Simocephalus from China (Crustacea: Diplostraca: Daphniidae). Acta Zootaxonomica Sinica. 19:403–411.
- Shi XL, Shi XB. 1996. On the species and distribution of Simocephalus in Heilongjiang Province, China (Branchiopoda: Diplostraca). Acta Zootaxonomica Sinica. 21:263–276.
- Xiang XF, Ji GH, Chen SZ, Yu GL, Xu L, Han BP, Kotov AA, Dumont HJ. 2015. Annotated Checklist of Chinese Cladocera (Crustacea: Branchiopoda). Part I. Haplopoda, Ctenopoda, Onychopoda and Anomopoda (families Daphniidae, Moinidae, Bosminidae, Ilyocryptidae). Zootaxa. 3904(1):1–27.