Abstract
Gentiana arethusae Burkill is a perennial herb classified in the Gentianaceae. In this study, the complete chloroplast genome of G. arethusae was sequenced and analyzed. The chloroplast genome is 137,458 bp in length and encodes a total of 116 genes, including 71 protein-coding, 37 tRNA, and eight rRNA genes. The genome has a low GC content of 38.0%. Phylogenetic analysis of the genome of G. arethusae resolved it in a clade with Gentiana obconica and Gentiana veitchiorum. The complete chloroplast genome of G. arethusae will be helpful to study the genetic diversity and phylogenetics of the Gentianaceae.
Gentiana arethusae Burkill is a perennial herb with a high ornamental value, classified in the Gentianaceae. The species is mainly distributed in Yunnan, Hubei, Sichuan, and Tibet provinces in China (Flora of China Editorial Committee Citation1988). Gentiana arethusae generally grows on hillsides and grasslands at an altitude of 2000–3000 m (Flora of China Editorial Committee Citation1988). Many species of the genus can be used in Chinese medicine, including G. arethusae which is used to treat rheumatic arthritis (Qing et al. Citation2014). It is well-established that the chloroplast (cp) genome provides effective information for the identification, phylogenetic relationships, and population genetic analysis of plant species (Shang et al. Citation2020). In previous research, the evolutionary history and phylogenetic relationship of G. arethusae were generally studied using ITS and TrnL-F sequences (Favre et al. Citation2014, Citation2016), however, the complete cp genome of G. arethusae has not yet been deciphered. Here, the cp genome of G. arethusae was sequenced, assembled and analyzed, and will be contributed to the conservation, genetic improvement, and sustainable management of this species.
A wild individual of G. arethusae was sampled from Wantan town (110°23′E, 28°56′N, 2020 m), Wufeng County in Hubei province of China. The voucher specimen was deposited at the Herbarium of Yunnan Agricultural University (Contact person: Hongzhi Wu, [email protected], No. 2021CD001). Total genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987). The genome sequence was performed using Illumina Novaseq PE150. The analysis yielded 3.61 Gb of raw data that was further trimmed and assembled using SPAdes (Bankevich et al. Citation2012). The annotation of the cp genome was conducted with Geneious 8.0.2 (Kearse et al. Citation2012). The annotated genomic sequence was submitted to GenBank under accession number MZ603883.
The complete cp genome of G. arethusae is 137,458 bp in length and consists of two inverted repeats 23,865 bp, separated by a large single-copy region (LSC 77,907 bp) and a small single-copy region (SSC 11,822 bp). The overall GC content is 38.0%, with the GC content of the LSC at 35.6%, and the GC content of the SSC at 30.3%. The cp genome contains a total of 116 functional genes, including 71 protein-coding, 37 tRNA, and eight rRNA genes. Among them, 12 distinct genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, atpF, rpoC1, petB, petD, rpl16, and rpl2) contain one intron and three genes (rps12, ycf3, and clpP) contain two introns. All the rRNA genes in the genome sequence were located in the repeat region. Furthermore, the cp genome of G. arethusae was similar in gene structure and arrangement to the previously published Gentian species (Sun et al. Citation2018).
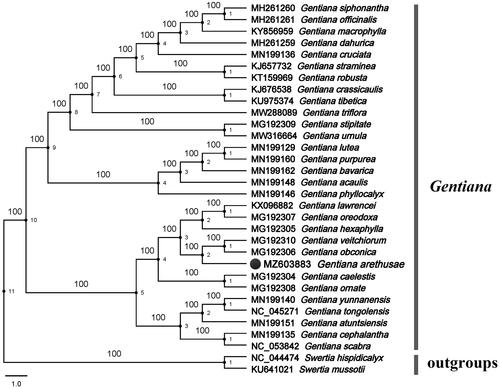
We used the complete cp genomes of G. arethusae and 29 other species from the genus Gentiana to reconstruct the phylogenetic tree designating Swertia mussotii and Swertia hispidicalyx as the outgroups. The maximum-likelihood (ML) analysis using RAxML 8.0 software (Stamatakis Citation2014) and the GTR + T nucleotide substitution model with 1000 bootstrap replicates resolved G. arethusae in a clade with Gentiana veitchiorum and G. obconica with high bootstrap values (). Previous researchers used a combination of nuclear (ITS) and plastid (trnL-F and atpB-rbcL) markers to study the phylogenetic relationship of Gentianinae, the results showed that G. arethusae was closely related to Gentiana lawrencei and Gentiana purdomiii, while G. veitchiorum was relatively distant (Favre et al. Citation2014). However, in our research, the phylogenetic relationship between G. arethusae and G. lawrencei is rather distant (the complete cp genome of G. purdomiii has not yet been deciphered), and more close to G. veitchiorum (). Some studies have shown that different conclusions are reached based on different molecular evidence for the same group (Zhang and Li Citation2011). The cp genome contains more nucleotide and amino acid information than gene sequences, and cp genomes are sequenced and analyzed more quickly and easily (Zhang and Li Citation2011). Therefore, the complete cp genome of G. arethusae will be helpful to determine the phylogenetic and evolutionary history of Gentiana.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MZ603883. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA749867, SRR15256470, and SAMN20423158, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Favre A, Matuszak S, Sun H, Liu E, Yuan YM, Muellner AN. 2014. Two new genera of Gentianinae (Gentianaceae): Sinogentiana and Kuepferia supported by molecular phylogenetic evidence. Taxon. 63(2):342–354.
- Favre A, Michalak I, Chen C-H, Wang J-C, Pringle JS, Matuszak S, Sun H, Yuan Y-M, Struwe L, Muellner-Riehl AN. 2016. Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J Biogeogr. 43(10):1967–1978.
- Flora of China Editorial Committee. 1988. Flora of China, Vol. 62. Beijing: Science Press; p. 148.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Qing SY, Lu YQ, Pang R, Lui DQ, Wang CH, Lui HK, Lu QG. 2014. Present situation investigation and development strategy of folk medicine in Wuxi county. Chinese J Ethnomed Ethnopharmacy. 23(09):22–23.
- Shang MY, Xiang XM, Fu LY, Jun Q, Bao ZD, Ying W. 2020. Assembly and phylogenetic analysis of the complete chloroplast genome sequence of Gentiana scabra Bunge. Mitochondrial DNA B. 5(2):1691–1692.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun S-S, Fu P-C, Zhou X-J, Cheng Y-W, Zhang F-Q, Chen S-L, Gao Q-B. 2018. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): insights into plastid gene loss and molecular evolution. Front Plant Sci. 9:493.
- Zhang YJ, Li DZ. 2011. Advances in phylogenomics based on complete chloroplast genomes. Plant Divers Resour. 33(4):365–375.