Abstract
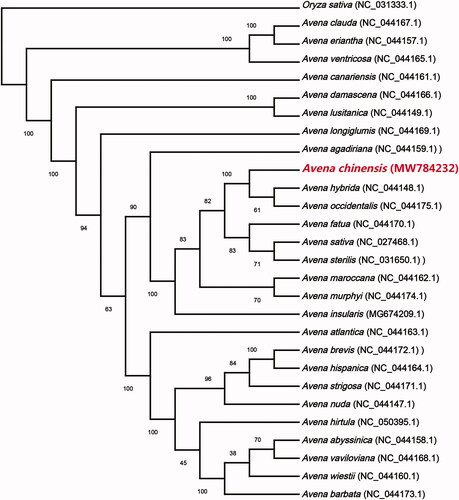
Avena chinensis is recognized as one of the cereals with high nutritional value in the world. In this study, the complete chloroplast (cp) genome sequence of A. chinensis was reported. The complete cp genome of A. chinensis was 135,899 bp in length with a GC content of 38.51%, including a large single copy (LSC) region of 80,117 bp, a small single copy (SSC) region of 12,576 bp, and a pair of inverted repeated regions of 21,603 bp. The A. chinensis cp genome encoded 128 functional genes, including 82 protein-coding genes, 38 tRNAs, and eight rRNAs. The phylogenetic analysis showed that A. chinensis was closely related to Avena hybrid and Avena occidentalis.
Avena chinensis (Fisch. ex Roem. & Schult.) Metzg. 1824, an annual herb of the genus Avena (Poaceae), is one of the most widely grown cereals in the world and a valuable resource in some countries, both for human consumption and animal feed (Fu et al. Citation2019). It is mainly distributed in the north, northwest and southwest of China in high latitude, high altitude, alpine arid and semi-arid areas due to its cold-loving, poor and drought-resistant characteristics (Liu et al. Citation2021). A. chinensis belonging to a tribe (Aveneae), separates from the other small-grained cereals such as wheat, barley, rye, triticale (Triticeae) and rice (Oryzeae), which contains 42 chromosomes, representing three distinct sets of nuclear genomes (A, C, and D) (Marshall et al. Citation2013; Yan et al. Citation2016). The protein and fat contents of A. chinensis are higher than husked oats, nevertheless fiber content is lower (Givens et al. Citation2004; Biel et al. Citation2009). In the present study, the complete chloroplast (cp) genome of A. chinensis (GenBank accession number: MW784232) was assembled to provide genomic and genetic sources for further research.
The fresh leaves of A. chinensis were collected from Huangzhong (101°31′ E, 36°28′ N), Qinghai Province, China. Total genomic DNA of A. chinensis was extracted from fresh leaves using the modified CTAB method and quantified (Allen et al. Citation2006). The voucher specimen and extracted DNA were deposited in the Herbarium of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (Handong Wang, [email protected]) under the voucher number WHD2020001. Genome sequencing was performed using the Illumina HiSeq Platform (Illumina, San Diego, CA) at Genepioneer Biotechnologies Inc. (Nanjing, China). Approximately, 26.12 million 150 bp paired-end reads were obtained, and 7.69 GB of clean data was generated after filtering. Then, the clean reads were assembled using SPAdes Version 3.10.1 (Bankevich et al. Citation2012), and the reference cp genome of Avena occidentalis (GenBank accession number: NC_044175.1) was used for quality control after assembly. Finally, the assembled genome was annotated in CpGAVAS (Liu et al. Citation2012).
The complete cp genome of A. chinensis was 135,899 bp in length with a GC content of 38.51%, including a large single copy (LSC) region of 80,117 bp, a small single copy (SSC) region of 12,576 bp, and a pair of inverted repeated regions of 21,603 bp. The A. chinensis cp genome encoded 128 functional genes, including 82 protein-coding genes, 38 tRNAs, and eight rRNAs.
The maximum-likelihood phylogenetic tree (ML tree) was generated based on the complete cp genome of A. chinensis and 26 other species of the genus Avena, with Oryza sativa as outgroup, of which the 27 cp genomes for phylogenetic analysis were downloaded from NCBI database. The 28 complete cp genome sequences were aligned by MAFFT v7.037 (Katoh and Standley Citation2013). The phylogenetic tree was built using MEGA X (Kumar et al. Citation2018) with bootstrap set to 1000. The phylogenetic tree showed that A. chinensis was closely related to Avena hybrid and Avena occidentalis (). This study was the first report on the complete cp genome of A. chinensis which could be useful for the phylogenetic and evolutionary studies of Avena and Poaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW784232. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA743926, SRR15044535, and SAMN20063186, respectively.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Biel W, Bobko K, Maciorowski R. 2009. Chemical composition and nutritive value of husked and naked oats grain. J Cereal Sci. 49(3):413–418.
- Fu YB, Li P, Biligetu B. 2019. Developing chloroplast genomic resources from 25 Avena species for the characterization of oat wild relative germplasm. Plants. 8(11):438.
- Givens DI, Davies TW, Laverick RM. 2004. Effect of variety, nitrogen fertiliser and various agronomic factors on the nutritive value of husked and naked oats grain. Anim Feed Sci Technol. 113(1–4):169–181.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Liu JX, Liu RR, Jia HY, Bu T, Li N. 2021. Physiological mechanism of NaHS priming improvement of seed vigor in naked oat. Acta Pratacult Sin. 30(2):135–142.
- Marshall A, Cowan S, Edwards S, Griffiths I, Howarth C, Langdon T, White E. 2013. Crops that feed the world 9. Oats—a cereal crop for human and livestock feed with industrial applications. Food Sec. 5(1):13–33.
- Yan HH, Bekele WA, Wight CP, Peng YY, Langdon T, Latta RG, Fu YB, Diederichsen A, Howarth CJ, Jellen EN, et al. 2016. High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat. Theor Appl Genet. 129(11):2133–2149.